Knowledge Integration in Precision Medicing
The research group 'Knowledge Integration in Precision Medicine' (KIP) at the UKE focuses on evaluating existing knowledge networks and IT infrastructures to optimize and reshape clinical and scientific processes. Our mission is also to advance the use of artificial intelligence (AI), especially Large Language Models (LLMs), in medicine to simplify data visualizations and analyses for medical researchers. The primary focus is often – but not exclusively – on oncological diseases, given that the group leader, Dr. Riemann, as a Ph.D. medical physicist, has extensive research experience in this field.
The team is, for instance, working on standardizing, simplifying, and expediting molecular tumor boards and the necessary literature review to provide more patients for whom standard oncological therapy has failed access to personalized treatment options. Additionally, the group is developing an innovative, privacy-compliant platform for hypothesis generation using secondary data, offering researchers straightforward access to pseudonymized data and comprehensive analysis capabilities.
Our interdisciplinary team collaborates closely with both internal clinical partners and national consortial projects. Together, we aim to develop innovative solutions for the multifaceted challenges of modern medical research, particularly in the context of precision medicine.
Current projects
-
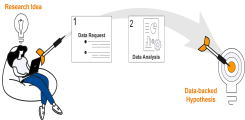
Data protection-compliant and uncomplicated testing of research hypotheses: the data hotel
Data protection-compliant and uncomplicated testing of research hypotheses: the data hotel
The data hotel is a UKE-wide platform established by the Institute for Applied Medical Informatics in October 2022 that enables data protection-compliant research and hypothesis generation based on secondary clinical data (Soarian, laboratory values, ICM). The data hotel enables clinical researchers to:
- Perform a cohort specification with data from 2008 to the present,
- Receive the pseudonymized patient data in a protected research environment (the 'hotel room'), and
- Analyze the data within the Jupyter environment using the programming languages Python, R, or Julia.
-
Acceleration and facilitation of molecular tumor boards: MONOCLE
Acceleration and facilitation of molecular tumor boards: MONOCLE
If oncological guideline therapy fails, molecular diagnostics are increasingly being carried out to provide the patient with a personalized treatment option. This process is very time-consuming and has not yet been standardized nationally. The aim is therefore to accelerate, simplify, and standardize the molecular tumor board (MTB) through (partial) automation, as well as the implementation of a standardized data model, the core data set of the German Network for Personalized Medicine (DNPM). The MOlecular ONcOlogy Clinical Evaluation (MONOCLE) web application was developed at our institute to map the processes, including both clinical and genetic information across different specialist areas, in a standardized way. MONOCLE helps with documentation and will also be able to access the Knowledge Connector (KC) tool developed by the DKFZ, which significantly speeds up the lengthy literature search for a personalized treatment approach.
Due to the modular structure of MONOCLE, it can and will be used for future projects in which structured reporting of certain clinical and genetic parameters is also required, e.g. nNGM or §64e. Research is also being conducted into the expansion of the KC with the help of Large Language Models (LLMs) concerning the automatic updating and revision of knowledge entries.
-
KiMeKo – AI-Med-Collaboration platform
KiMeKo – AI-Med-Collaboration platform
KiMeKo aims to establish a collaborative AI-driven medical platform for northern Germany. Building on previous research and development initiatives in AI-based medical applications, the platform will provide methods, tools, workflows, and organizational support to streamline the development of AI solutions in healthcare.
The AI-Med ecosystem will offer standardized services, methodologies, and workflows wherever possible. By fostering seamless collaboration between medical professionals, computer scientists, and industry partners, the platform aims to significantly enhance both the speed and quality of AI-driven medical innovations. This will increase the feasibility of large-scale, strategic AI applications with broad potential for improving healthcare delivery.
For more information please go to the official KiMeKo Website.
Publications
2025
Macromolecule Modelling for Improved Metabolite Quantification Using Short Echo Time Brain 1H-MRS at 3 T and 7 T: The PRaMM Model
Dell'Orco A, Riemann L, Ellison S, Aydin S, Göschel L, Ittermann B, Tietze A, Scheel M, Fillmer A
NMR BIOMED. 2025;38(1):e5299.
2024
Plasma p-tau181 and GFAP reflect 7T MR-derived changes in Alzheimer's disease: A longitudinal study of structural and functional MRI and MRS
Göschel L, Dell'Orco A, Fillmer A, Aydin S, Ittermann B, Riemann L, Lehmann S, Cano S, Melin J, Pendrill L, Hoede P, Teunissen C, Schwarz C, Grittner U, Körtvélyessy P, Flöel A
ALZHEIMERS DEMENT. 2024;20(12):8684-8699.
An LLM-Based Visualization and Analysis Aid for a Secondary Use Clinical Data Platform
Spiegel S, Wendt T, Gundler C, Ückert F, Riemann L
Stud Health Technol Inform. 2024;316:1617-1621.
2023
7T amygdala and hippocampus subfields in volumetry-based associations with memory: A 3-year follow-up study of early Alzheimer’s disease
Göschel L, Kurz L, Dell'Orco A, Köbe T, Körtvelyessy P, Fillmer A, Aydin S, Riemann L, Wang H, Ittermann B, Grittner U, Flöel A
NEUROIMAGE-CLIN. 2023;38:103439.
An Open-Source Software Tool to Facilitate Data Protection Impact Assessments
Riemann L, Hähner F, Schmitz A, Ataian M, Jaster M, Ückert F
APPL SCI-BASEL. 2023;13(20):.
Introduction of MONOCLE – a software to reduce the workload and optimize the processes of the molecular tumor board at the University Hospital Hamburg-Eppendorf
Schmitz A, Lauk K, Heß K, Voß H, Fulla O, Schönbeck N, Ückert F, Gundler C, Riemann L
Ger Med Sci. 2023.
Letzte Aktualisierung aus dem FIS: 16.04.2025 - 04:20 Uhr