iDfellows
-
Patricia Bartsch (1. CS)
Staphylococcus epidermidis innate immune evasion during implant infections
Project leader: Prof. Dr. Holger Rohde
Affiliation: Institute for Medical Microbiology, Virology and Hygiene
Background and preliminary data: Staphylococcus epidermidis is a leading pathogen in implant-associated infections. Biofilm formation protects S. epidermidis from innate immune effector mechanisms, and plays a key role for persistence in in a hostile environment. The underlying molecular mechanisms are not yet understood. Previous work from our lab has provided evidence that biofilm formation interferes with phagocytosis, bacterial killing and pro-inflammatory macrophage activation. More recently, using S. epidermidis wild-type strains and specific mutants in combination with live cell imaging approaches, evidence was obtained that S. epidermidis is able survive within primary human macrophages. While an isogenic biofilm-negative mutant was readily degraded and almost completely killed with 12 hours after up-take, the biofilm forming S. epidermidis wild type strain was able to persist within macrophages for up to 72 hours. So far, it is a common opinion that S. epidermidis is a strictly extracellular pathogen and lacks the ability to survive intracellularly. Therefore, our finding add an important novel and so far unrecognized piece of evidence to our picture of S. epidermidis pathogenicity and the pathogenesis of chronic implant infections.
Hypothesis: Building on own preliminary observations we hypothesize that S. epidermidis employs defined mechanisms that facilitate intracellular pathogen survival in macrophages.
Aims and Work Program: 1. Dissection of molecular events during up-take and intracellular fate of S. epidermidis in human macrophages. 2. Identification of S. epidermidis factors relevant to within macrophage survival. 3. Importance of intracellular survival for the pathogensis of implant infections.
Aim #1 Work package 1 aims at dissecting the molecular events and cellular pathways contributing to S. epidermidis phagocytosis. Using a recently developed model to study macrophage - biofilm interactions, to this end S. epidermidis wildtype 1457 and a series of fluorescence labeled derivatives will be employed in live cell imaging experiments focusing on the very early phase of pathogen-macrophage contact (0 - 20 h). To specifically characterize the contribution of biofilm forming ability, a biofilm-negative mutant shall be used as a comparator. Imaging and quantification of S. epidermidis uptake as well as identification of cellular compartments involved in pathogen handling will be substantially aided by the close collaboration with the UKE microscope imaging facility Umif and infrastructure from RTG Humans and Microbes (M. Aepfelbacher, Stefan Linder, Thomas Braulke). Making use of their established methods, a major goal will be to draw a detailed functional picture of relevant host cell events during S. epidermidis up-take. In extension to imaging driven analysis, transcriptomics shall be employed to more precisely define host pathways engaged by biofilm forming S. epidermidis. Importantly, by parallel approaching host and pathogen transcriptomes using a dualSeq strategy, macrophage responses will be linked to adaptive transcriptional S. epidermidis events. Transcriptomics essentially depend on well-established collaborations with in-house bioinformatics resources (Jiabin Huang) and infrastructures from the SFB 1192 (Christian Krebs). Using biofilm-negative mutant 1457-M10, the contribution of biofilm forming ability to evade macrophage clearance can be estimated.
In Aim #2 Preliminary evidence supports the idea that S. epidermidis engages defined mechanisms to persist in macrophages. A two-tier approach shall be employed to identify those functionally involved pathogen factors. Firstly, a high density Bursa aurealis Tn-mutant library of S. epidermidis 1457 will be established. By testing the library in a macrophage infection model and a high-throughput FACS assay to quantify intracellular S. epidermidis, mutations will be selected that are associated with decreased intracellular S. epidermidis numbers. Results will feed into experiments in which specific mutations of identified genes are being generated using a recently developed CRISPRr/Cpf1 based genome editing system. CRISPR/Cpf1-based genome editing is also used in the second complementary approach to Tn mutagenesis. Making use of results from dualSeq experiments, genes regulated after macrophage contact will be inactivated. Mutants from Tn mutagenesis and CRISPR/Cpf editing will then be tested using imaging approaches as used in WP1 to nail down their specific contribution for S. epidermidis handling by macrophages.
In Aim #3, So far, evidence for intracellular S. epidermidis survival of S. epidermidis is limited to in vitro experiments. Currently, our group is involved in establishment of a mouse implant-infection model (collaboration with Johannes Keller, experimentelle Unfallchirurgie). The model shall be used to quantify survival of S. epidermidis wildtype 1457 and corresponding biofilm-negative mutant 1457-M10. Relying on available antisera to stain for S. epidermidis, ex vivo material will be investigated with a special focus on characterizing and enumerating intra- and extracellular bacteria. Similarly, we will make use of human ex vivo material from PJI (collaboration Helios ENDO-Klinik) to characterize the presence of intracellular S. epidermidis using microscopic and FACS approaches. Finally, the infection model will serve to test the in vivo relevance of S. epidermidis genes identified in aim 2 for pathogenesis of S. epidermidis implant infections.
Project-related publications:
1. Stamm J, Weißelberg S, Both A, Failla AV, Nordholt G, Büttner H, Linder S, Aepfelbacher M, Rohde H. 2022. Development of an artificial synovial fluid useful for studying Staphylococcus epidermidis joint infections. Front Cell Infect Microbiol. 2022 Jul 29;12:948151. doi: 10.3389/fcimb.2022.948151. eCollection 2022.
2. Bartsch P, Kilian C, Hellmig M, Paust HJ, Borchers A, Sivayoganathan A, Enk L, Zhao Y, Shaikh N, Büttner H, Wong MN, Puelles VG, Wiech T, Flavell R, Huber TB, Turner JE, Bonn S, Huber S, Gagliani N, Mittrücker HW, Rohde H, Panzer U, Krebs CF.. 2022. Th17 cell plasticity towards a T-bet-dependent Th1 phenotype is required for bacterial control in Staphylococcus aureus infection. PLoS Pathog. 2022 Apr 21;18(4):e1010430. doi: 10.1371/journal.ppat.1010430. eCollection 2022 Apr.
3. Both A, Huang J, Qi M, Lausmann C, Weißelberg S, Büttner H, Lezius S, Failla AV, Christner M, Stegger M, Gehrke T, Baig S, Citak M, Alawi M, Aepfelbacher M, Rohde H. 2021. Distinct clonal lineages and within-host diversification shape invasive Staphylococcus epidermidis populations PLoS Pathog. 2021 Feb 5;17(2):e1009304. doi: 10.1371/journal.ppat.1009304. eCollection 2021 Feb. PMID: 33544760
4. Du X, Larsen J, Li M, Walter A, Slavetinsky C, Both A, Sanchez Carballo PM, Stegger M, Lehmann E, Liu Y, Liu J, Slavetinsky J, Duda KA, Krismer B, Heilbronner S, Weidenmaier C, Mayer C, Rohde H, Winstel V, Peschel A.. 2021. Staphylococcus epidermidis clones express Staphylococcus aureus-type wall teichoic acid to shift from a commensal to pathogen lifestyle. Nat Microbiol. 2021 Jun;6(6):757-768. doi: 10.1038/s41564-021-00913
5. Büttner H, Perbandt M, Kohler T, Kikhney A, Wolters M, Christner C, Heise M, Wilde J, Weißelberg S, Both A, Betzel C, Hammerschmidt S, Svergun D, Aepfelbacher M, Rohde H. 2020. A giant extracellular matrix binding protein of Staphylococcus epidermidis binds surface-immobilized fibronectin via a novel mechanism. mBio. 2020 Oct 20;11(5):e01612-20. doi: 10.1128/mBio.01612-20
-
Christoph Kilian (2. CS)
Analysis of the role of CMV-reactive tissue-resident T cells on intestinal inflammation in IBD
Project leader: Prof. Dr. Samuel Huber
Affiliation: I. Department of Medicine, University Medical Center Hamburg-Eppendorf
Background and preliminary data:
Inflammatory bowel diseases (IBD) are immune-mediated diseases characterized by recurrent inflammation of the intestinal tract. The etiology of those chronic disorders is not completely understood. It is postulated that lifestyle factors and the microbiota can lead to a dysregulation of the mucosal immune system in genetically susceptible individuals. Interestingly, clinical studies suggest that viral infections can also affect the disease course in IBD patients. Compared to healthy individuals, IBD patients have a higher risk of the development of CMV (cytomegalovirus) colitis. Interestingly, CMV colitis is also associated with disease activity and steroid resistance in patients with ulcerative colitis. Indeed, infectious disease are known to be able to precede and aggravate various autoimmune and immune-mediated diseases. While the mechanisms underlying this phenomenon are largely unknown, several studies suggest that the induction of tissue resident memory T cells (TRM) might play a crucial role in this setting. These cells are categorized as a distinct memory T cell subset that usually does not take part in cell migration but adheres to a specific tissue. TRM cells can occur after infection and are considered to provide a protective function against reinfection. Phenotypically, they are characterized by the expression of high levels of CD69 and low levels of S1PR1. Interestingly, recent publications have shown that the induction of TRM cells upon infection with pathogens can aggravate the subsequent course of immune-mediated diseases even after clearance of the initial pathogen.
Based on these findings, we aim to test whether CMV infection can affect the course of IBD by inducing tissue-resident T cells in the intestine.
Hypothesis:
We hypothesize that CMV induces tissue-resident memory cells in the intestine and thereby contributes to an exacerbated intestinal inflammation during in IBD.
Aims and Work Programme:
We want to test this hypothesis using a translational approach and by studying both IBD/CMV colitis in IBD patients and by using murine CMV (MCMV) as a mouse model for CMV infection. Specifically, we want to pursue the following aims:
- Aim #1: Study of the immune cell composition and the clonal expansion of tissue-resident memory T cells in patients with IBD/CMV colitis
- Aim #2 Using a murine model for CMV Infection (MCMV), we aim to test whether there is a causal link between MCMV infection and a dysregulated mucosal immune response, which in turn leads to an exacerbation of colitis.
In Aim #1, we plan to collect intestinal biopsies from IBD patients with active colitis. Specifically, our aim is to compare patients with active IBD and CMV colitis (determined by positive CMV PCR in the biopsy specimens) compared to active IBD without CMV colitis. To this end, we will compare the immune cell composition in those two groups using flow cytometry and our staining panel will be focused on the in depth-characterization of effector and memory T cells. Furthermore, we plan to perform single-cell sequencing and TCR-sequencing of isolated mucosal T cells. Based on ligand-receptor interactions, we can thus identify CMV-reactive T cells in the mucosa of patients with active IBD vs. IBD/CMV colitis. Furthermore, we will be able to monitor, whether a clonal expansion of CMV-reactive T cells can be observed in this setting and whether those clonally expanding cells indeed have tissue-resident characteristics. Finally, we will correlate our flow cytometry results with our single-cell sequencing data in order to test, whether those clonally-expanding, tissue-resident CMV-reactive T cells are indeed contributing to a strongly dysregulated immune response in IBD/CMV colitis. Those results will be incremental to understand the mechanism of why CMV colitis is associated with a complicated disease course in IBD patients.
In Aim #2, we aim to test the hypothesis that latent MCMV infection leads to a dysregulated mucosal T cell-composition, which in turn leads to an exacerbation of colitis. To test this, we aim to induce a latent MCMV infection in mice. In the next step, we will use different murine models for colitis (DSS-colitis as a model of chemically induced colitis and Il10-/- mice as a genetic colitis model) in order to test, whether latent MCMV infection leads to an exacerbation of colitis in experimental colitis models. The use of those animal models (together with the MCMV infection) has already been approved by the responsible authorities (Genehmigung eines Tierversuchsvorhabens nach § 8 Abs. 1 des Tierschutzgesetzes). Furthermore, we plan to perform an in-depth study of the mucosal immune cell composition in those murine models. Specifically, we aim to analyze the T cell composition by FACS and single-cell RNA as well as TCR sequencing, as described in Aim #1. This will allow us to determine, whether tissue-resident, MCMV-reactive T cells are clonally expanding after the induction of experimental colitis in mice. Since IBD (in the case of Crohn’s Disease) can also affect the small intestine, we aim to further test the role of latent CMV infection in a murine model of small intestinal inflammation that is already established in our lab (the aCD3-model of small intestinal inflammation which is induced by the injection of CD3-specific antibodies). This model would allow us to study the functional role of tissue-resident immune cells after CMV infection in more detail by performing small intestinal transplantation after the CMV infection. Our long-term collaborator Dr. Giannou has learned rodent small intestinal transplantation and would therefore enable us to study the role of CMV-reactive tissue-resident cells using a highly sophisticated state-of-the-art method. Overall, we believe that our animal studies can pave to the way towards a novel understanding of how viral infections can induce tissue-resident T cells which then play a causal role in controlling the course of immune-mediated disorders.
In conclusion, we believe that this project will offer novel insights into how CMV infection can induce tissue-resident intestinal T cells, which in turn exacerbate colitis in IBD patients. Understanding the mechanisms underlying this phenomenon could pave the way towards novel treatments for IBD.
Project-related publications:
1. Kempski J*, Giannou AD, Riecken K, Zhao L, …, Huber S. IL22BP Mediates the Antitumor Effects of Lymphotoxin Against Colorectal Tumors in Mice and Humans. Gastroenterology. 2020;159(4):1417-1430.e3.
2. Perez LG, Kempski J, McGee HM, Pelzcar P, …, Huber S. TGF-β signaling in Th17 cells promotes IL-22 production and colitis-associated colon cancer. Nat Commun. 2020;11(1):2608.
3. Brockmann L, Soukou S, Steglich B, Czarnewski P, …, Huber S. Molecular and functional heterogeneity of IL-10 producing CD4+ T cells. Nat Commun. 2018 May 25;11(1):2608; 9(1):5457
4. Pelczar P*, Witkowski M*, Perez LG*, Kempski J, …, Huber S. A pathogenic role for T cell-derived IL-22BP in inflammatory bowel disease. Science. 2016;354(6310):358-362.
-
Britta Zecher (3. CS)
Effect of KIR3DS1/3DL1 co-expression and copy number variation on anti-viral activity of NK cells
Project leader: Prof. Dr. Marcus Altfeld
Affiliation: Leibniz Institute of Virology, Hamburg
Background and preliminary data:
KIR3DS1 and KIR3DL1 belong to the family of killer cell immunoglobulin like receptors (KIRs) that play an important role in modulating NK cell function, including control of viral infections. Both KIR3DS1 and KIR3DL1 have been described to be involved in the control of viral infections such as human immunodeficiency virus (HIV)1, human adenovirus (hAdV)2, BK polyomavirus (BKV)3 and hepatitis C virus (HCV)4 infection. This protective effect has been observed in individuals expressing both KIR3DS1 and KIR3DL1, but not expressing KIR3DS1 alone. However, to date the mechanisms of NK cell mediated control of viral infection requiring both KIR3DS1 and KIR3DL1 remain unclear.KIRs are divided into two groups according to their function, which is linked to the intracytoplasmic domain. Activating KIRs have a short intracytoplasmic domain with an ITAM motif (KIRxDSx), while inhibitory KIRs have a long intracytoplasmic domain with an ITIM motif (KIRxDLx). Recognition of self-MHC class I molecules by inhibitory KIRs promotes tolerance towards endogenous cells in a process termed NK cell education. In consequence, downregulation of self MHC-class I molecules in malignant transformed or virus-infected cells leads to missing self-recognition by NK cells.KIR3DS1 and KIR3DL1, representing an activating and inhibitory receptor respectively, are encoded at the same locus and therefore represent alleles of one gene. Individuals can thus be homozygous or heterozygous for KIR3DS1 and KIR3DL1. Despite the high homology in the amino acid sequence, KIR3DS1 and KIR3DL1 have different ligands5. While KIR3DL1 binds to HLA-Bw4 molecules, the non-classical HLA-class I molecule HLA-F, that is upregulated e.g. on virus infected cells, serves as ligand for KIR3DS1 (Garcia-Beltran WF et al., Nat Immunol 2016). However, to date it is not clear whether KIR3DS1 and KIR3DL1 are co-expressed by the same NK cell on a single cell level, and how the expression pattern of KIR3DS1 and KIR3DL1 shapes the anti-viral activity of NK cells. Furthermore, the KIR3DS1/KIR3DL1 locus underlies copy number variations (CNVs). The total allele count for KIR3DS1/KIR3DL1 has been described to vary between one and three copies. In the third part of the project, we will therefore investigate the influence of CNV on KIR3DS1 and KIR3DL1 surface expression and subsequently the impact on anti-viral activity of the NK cells.
Hypothesis:
The co-expression of KIR3DS1 and KIR3DL1 influences the anti-viral activity of NK cells on a single cell level
Aims and Work Programme:
1. Determine (co-)expression of KIR3DS1/DL1 on a single cell level
2. Assess the functional impact of KIR3DS1/3DL1 co-expression on the anti-viral activity of NK cells
3. Modulation of anti-viral activity of NK cells through KIR3DS1/KIR3DL1 copy number variations
Work programme aim #1:
Healthy donors will be typed for HLA- and KIR alleles. Co-expression patterns of KIR3DS1/KIR3DL1 will be determined on NK cells of donors being heterozygous for these alleles. Representing two alleles of the same gene, KIR3DS1 and KIR3DL1 have a homology of >95% in their extracellular domain. Consequently, there are currently only KIR3DS1 monospecific and KIR3DS1/3DL1 bispecific antibodies available. Via flow cytometry, it is therefore only possible to distinguish between KIR3DS1-/KIR3DL1-, KIR3DS1+/KIR3DL1- andKIR3DL1+ cells that may or may not co-express KIR3DS1. To determine the co-expression pattern of KIR3DS1 and KIR3DL1, individual single NK cells of KIR3DS1/KIR3DL1 heterozygous, healthy individuals will be sorted via fluorescence activated cell sorting (FACS). Subsequently, the expression of KIR3DL1 and KIR3DS1 on an mRNA level will be determined via single-cell RT-qPCR (Fluidigm Technology).
Work programme aim #2:
To determine the functional impact of KIR3DS1/KIR3DL1 co-expression on the anti-viral activity of NK cells, NK cell clones from single cell sorted primary NK cells of healthy donors heterozygous for KIR3DS1 and KIR3DL1 will be generated. KIR3DS1 and KIR3DL1 expression of the clones will be determined via flow cytometry and RT-qPCR, as described above. The selected donors will be HLA-Bw4 positive or negative. This will allow us to differentiate between KIR3DL1/HLA-Bw4 educated or non-educated NK cells. NK cell clones expressing KIR3DS1 and/or KIR3DL1 of HLA-Bw4 positive or negative donors will be incubated with the respective ligands HLA-Bw4 and HLA-F (protein-coated beads and transfected cell lines). NK cell activation in will be determined by flow cytometric analysis of degranulation and cytokine expression.After dissecting the required KIR3DL1/KIR3DL1 expression pattern and genetic background for effective NK cell activation, the impact on control of viral infections will be assessed. We will use infection models that are well established in our lab and for which an influence of viral control by KIR3DS1 and KIR3DL1 has been demonstrated, such as HIV or hAdV infection. Antiviral activity of the NK cell clones will be determined via assessment of NK cell degranulation and killing of virus-infected cells, as well as inhibition of viral replication.
Work programme aim #3:
To assess anti-viral NK cell activity in the context of CNV, KIR3DS1/KIR3DL1 copy numbers will be determined via digital droplet PCR. NK cell clones will be generated from individuals with different copy numbers of KIR3DS1 and KIR3DL1, and the consequences for KI3DS1 and KIR3DL1 expression on the single cell level will be determined, as described in aim #1. The effect of CNV on the control of viral infection will be assessed, as described in aim #2.
Project-related publications:
1. Pelak K, Need AC, Fellay J, Shianna KV, Feng S, Urban TJ, Ge D, De Luca A, Martinez-Picado J, Wolinsky SM, Martinson JJ, Jamieson BD, Bream JH, Martin MP, Borrow P, Letvin NL, McMichael AJ, Haynes BF, Telenti A, Carrington M, Goldstein DB, Alter G; NIAID Center for HIV/AIDS Vaccine Immunology. Copy number variation of KIR genes influences HIV-1 control. PLoS Biol. 2011 Nov;9(11):e1001208.
2. Jung JM, Ching W, Baumdick ME, Hofmann-Sieber H, Bosse JB, Koyro T, Möller KJ, Wegner L, Niehrs A, Russu K, Ohms M, Zhang W, Ehrhardt A, Duisters K, Spierings E, Hölzemer A, Körner C, Jansen SA, Peine S, Königs I, Lütgehetmann M, Perez D, Reinshagen K, Lindemans CA, Altfeld M, Belderbos M, Dobner T, Bunders MJ. KIR3DS1 directs NK cell-mediated protection against human adenovirus infections. Sci Immunol. 2021 Sep 17
3. Koyro TF, Kraus E, Lunemann S, Hölzemer A, Wulf S, Jung J, Fittje P, Henseling F, Körner C, Huber TB, Grundhoff A, Wiech T, Panzer U, Fischer N, Altfeld M. Upregulation of HLA-F expression by BK polyomavirus infection induces immune recognition by KIR3DS1-positive natural killer cells. Kidney Int. 2021 May;99(5):1140-1148.
4. Lunemann S, Schöbel A, Kah J, Fittje P, Hölzemer A, Langeneckert AE, Hess LU, Poch T, Martrus G, Garcia-Beltran WF, Körner C, Ziegler AE, Richert L, Oldhafer KJ, Schulze Zur Wiesch J, Schramm C, Dandri M, Herker E, Altfeld M. Interactions Between KIR3DS1 and HLA-F Activate Natural Killer Cells to Control HCV Replication in Cell Culture. Gastroenterology. 2018 Nov;155(5):1366-1371.e3.
5. O'Connor GM, Guinan KJ, Cunningham RT, Middleton D, Parham P, Gardiner CM. Functional polymorphism of the KIR3DL1/S1 receptor on human NK cells. J Immunol. 2007 Jan 1;178(1):235-41.
-
Ilka Grewe (4. CS)
Dissecting the Role of T follicular helper cells in vaccination against Betacoronaviruses: Clonal evolution and TCR specificity
Project leader: Prof. Dr. med. Marylyn M. Addo, MSc, DTM&H
Affiliation: Director, Institute for Infection Research and Vaccine Development (IIRVD) Center for Internal Medicine (ZIM), UKE
Email: m.addo@uke.de, fon: 040-7410-51102
Background and preliminary data:
Emerging infectious diseases: Emerging infectious diseases (EID) represent a major threat to public health worldwide and were recently named among the top ten threats to global health by the World Health Organization (WHO). The Ebola virus disease (EVD) crisis in West Africa and the current coronavirus disease (COVID)-19 pandemic demonstrated that the world community was ill prepared and challenged by developing effective interventions for the control of severe EID outbreaks in a timely manner. In response to the current COVID-19 pandemic of historic scale, the speed of vaccine development against SARS-CoV-2 and implementation of clinical trials have been unprecedented. However critical knowledge gaps remain to be addressed to evaluate vaccine candidates at an early stage of vaccine development.
Challenges in the development of vaccines against emerging infectious diseases: Vaccines are among the most successful public health interventions. Current licensed vaccines have been mostly generated by empiric strategies and the exact mechanisms that mediate vaccine-induced protection remain inadequately understood. In light of strategic vaccine development against EID, there is a critical need to better understand how vaccines work and how correlates of vaccine-induced immunogenicity and protection can be assessed at an early stage (i) to facilitate decision-making on the next stages of vaccine development, (ii) to evaluate vulnerable cohorts (iii), and to improve the design of novel vaccine candidates. Phase I clinical trials in humans are thus a critical step in vaccine development as through intensive sampling and comprehensive analysis of study subjects, they allow for critical insight into both safety and immunogenicity of new vaccine constructs.
Vaccine-induced immunogenicity: Adaptive immunity induced by vaccines is critical for vaccine efficacy. While analysis of antigen-specific T and B cells as well as antibodies is a standard approach to evaluate vaccine-induced immunogenicity, T follicular helper (TFH) cells have been understudied to date. TFH cell populations play a key role in B cell selection and antibody affinity maturation during GC reactions and are essential for generation of protective B cell responses. Thus, induction of a strong antigen-specific TFH response upon immunization represents a key goal for the design of vaccines and may emerge as a predictive marker and/or a correlate for protection against infection or disease.
Preliminary work: Clinical trials to evaluate vaccine candidates against MERS and COVID-19: As part of the TTU Emerging Infections of the German Center for Infection Research (DZIF), our group has a specific expertise in early phase clinical testing of vaccine candidates against emerging viruses. We recently performed investigator-initiated (IIT) vaccine trials evaluating the viral-vector vaccine candidates VSV-EBOV, MVA-MERS-S and MVA-SARS-2-S/ST against Ebola virus disease, MERS and COVID-19, respectively. Multiple samples from healthy vaccine volunteers pre- and early post-vaccination were collected to gain comprehensive insights into innate and adaptive immune mechanisms induced by vaccination. Analysis include early gene expression profiles associated with the candidate vaccine, as well as humoral responses and detailed T and B cell analyses in response to vaccination.
Hypothesis: We hypothesize that early induction of TFH cells populations results in strong neutralizing and non-neutralizing humoral immunity. Detailed longitudinal analyses of TFH cell populations will provide critical insight into molecular and cellular mechanisms related to the kinetics of TFH cell populations and optimal induction of B cell help.
Aims and Work Programme: Human TFH cell populations will be comprehensively analyzed to understand mechanisms of antigen-delivery systems that confer protective immunity. We will take advantage of longitudinally collected human samples originating from the Phase I/Ib clinical trials testing MVA-MERS-S and COVID-19 vaccine studies. Longitudinal samples from SARS-CoV-2-positive individuals will be included as controls.
Figure 1. In-depth analysis of TFH cell populations following betacoronavirus vaccination or infection. Longitudinal sample collection in combination with different technologies (Luminex, Flowcytometer, TCR Sequencing) to evaluate TFH dynamics on cellular and molecular level. All data sets will be analyzed using R and/or Prism.
WP 1: Profiling of antigen-specific TFH cell populations following natural infection and vaccination. Characterize TFH cell responses to MERS-CoV/SARS-CoV-2 spike, analyzed from participants of different vaccine trials and compare responses with samples from infected individuals. Classical assays based on flow cytometry and Luminex/Legendplex will be applied to identify TFH1, TFH2 and TFH17 cell populations and their activation status. The aim is to longitudinally investigate TFH cell populations induced by different vaccine candidates against coronaviruses (MVA-based vaccines, mRNA-based vaccines), and to compare data between vaccinees and infected individuals.
WP 2: TCR repertoire analysis of TFH cell population. A subgroup of each cohort will be included in the analysis of the TCR gene. Flowcytometric cell sorting will be applied to isolate PD1highICOS+ TFH cells from specific timepoints following vaccination or infection. The aim is to sequence the TRB and TRA genes and to investigate the clonal expansion of TFH cells following immunization or infection.
WP3:Comparison of TFH cell signatures with antibody responses induced by vaccine candidates or infection. Our cohorts were comprehensively analyzed for humoral responses. These data will be used to associate TFH cell populations with antibody responses.
Project-related publications:
1. Agnandji ST#, Huttner A#, Zinser ME#, Njuguna P#, Dahlke C, …., Lohse AW, …, Ramharter M, …, Bejon P#, Kremsner PG#, Siegrist CA#, Addo MM#. Phase I Trials of rVSV Ebola Vaccine in Africa and Europe. N Engl J Med 2016 374:1647-60
2. Dahlke C#, Kasonta R#, Lunemann S#, …, Lohse AW, Becker S, Addo MM; VEBCON Consortium. Dose-dependent T-cell Dynamics and Cytokine Cascade Following rVSV-ZEBOV Immunization. EBioMedicine 2017;19:107-18.
3. Koch T#, Dahlke C#, Fathi A, …, Sutter G, Becker S, Addo MM. Safety and immunogenicity of a modified vaccinia virus Ankara vector vaccine candidate for Middle East respiratory syndrome: an open-label, phase 1 trial. Lancet Infect Dis 2020;20:827-38.
4. Weskamm M, …., Dahlke C#, Addo MM#. Persistence of MERS-CoV-spike-specific B cells and antibodies after late third immunization with the MVA-MERS-S vaccine. Cell Rep Med 2022, Jul 19;3(7):100685
5. Fathi A, Dahlke C, …, Haagmans BL, Sutter G, Becker S, Addo MM. Increased neutralization and IgG epitope identification after MVA-MERS-S booster vaccination against Middle East respiratory syndrome. Nat Commun 2022 Jul 19;13(1):4182. #equally contributing authors
-
Aenne Harberts (5. CS)
Trajectories of immune dysfunction between the liver, the intestine andluminal flora in advanced chronic liver disease
Project leader: PD Dr. med. Peter Huebener
Affiliation: Department of Medicine, Ist Medical Clinic and Polyclinic, University MedicalCenter Hamburg-Eppendorf
Background and preliminary data:
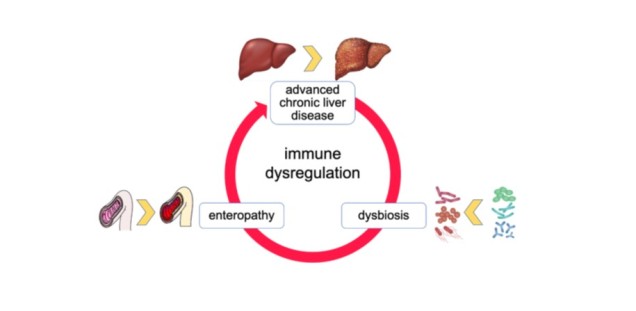
Prevalences of chronic liver diseases areincreasing worldwide, and infections aremajor drivers of morbidity and mortality inthis patient cohort. A dysfunctionalinterplay between the liver, the intestinalmucosa and the luminal microbiota -three anatomically and functionallyclosely intertwined biological systems - isbelieved to favor infections, but also toprovide aims for prophylactic andtherapeutic medical interventions (1).
Here, we aim to determine dysfunctional trajectories of these three biological entities duringinfections, and to characterize their therapeutic accessibility. We plan 3 major project goals includinganimal experiments, clinical observations as well as interventional studies in human subjects.
Hypothesis:
Dysregulated immune responses to infections in advanced chronic liver diseases (ACLD) are driven by defective inner- and inter-system crosstalk between the liver, the gut and theintestinal microbiota, which may be exploited for prophylactic or therapeutic interventions.
Aims and Work Programme:
Aim 1: Patterns of pathogen-induced immune dysregulation in ACLD: We aim for a comprehensive phenotypic and molecular characterization of immune dysregulation asa function of progressive liver diseases.
In Aim 1A) of the proposal, the effects of well-controlled infectious stimuli are to be examined againstthe background of pathogenetically diverse and differently advanced liver diseases in rodents. Twomouse models of chronic liver damage are primarily used:
• the Tak1Δhep model of chronic progressive cell death-dependent hepatopathy (2)
• the Mdr2-/- model of progressive cholestatic liver disease
By using two genetic models of liver damage, hepatopathy-specific and overarching mechanisms ofimmune dysfunction in liver diseases can be identified. Experimental animals (and controls) areexposed to bacterial pathogens (Str. pneumoniae, E. coli) systemically (i.v.) or locally (i.t., i.p.) atdifferent stages of liver disease. Target variables are a) cellular compositions of the liver, spleen,intestine, bone marrow, regional lymph nodes and circulating blood, b) biochemical analyses of theimmune response including multiplex ELISA, bulk and single cell-sequencing, Cytof-analyses,immunostaining), c) infection-associated alterations of the intestinal flora, permeability of theintestinal wall and epithelial cell death processes, and are supplemented by d) in vitro experimentalsetups of primary cell cultures from the corresponding experimental animals.
Aim 2: Intestinal dysbiosis as a modifiable effector system of ACLD-immune dysregulation. ACLD isassociated with characteristic alterations of the intestinal flora, whose pathophysiological roles areunclear. Here, we aim to characterize the consequences of intestinal dysbiosis on 1) intestinalinflammation and permeability, 2) systemic immune responses to acute infections, and 3) theprogression of liver disease.
Part (2A) The influence of chronic liver diseases will be characterized using stool samples from testanimals from application part 1A).
Animals: Tak1Δhep and Tak1f/f per n=10 mice aged 4 weeks, 16 weeks and 48 weeks, respectively.Samples from the deep small intestine, proximal colon and stool pellets as well as from test animalswith infections (see part 1A of the application) before/after targeted antibiotic therapies.The Tak1Δhep mouse samples offer - since the genetic defect is limited to the liver - the advantageof a longitudinal observation of microbial flora alterations that may exacerbate with progressive liverdisease and are otherwise largely free of confounding factors - even in regions that are difficult toaccess in patients, i.e. the small intestine. Separate housing of the animals prevents homogenizationof the microbiota through coprophagy. The focus of the analyses is on the composition of theintestinal flora by 16S sequencing and shotgun metagenomics on selected, representative samples.
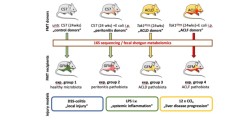
In sub-project 2B), potential pathogenetic properties of intestinal dysbiosis are characterized. Usingfecal microbiome transfer (FMT), the microbiome of ACLD, septic or ACLF experimental animalsfrom Aim 1A) are transplanted into germ-free experimental animals. The pathophysiologicalconsequences of intestinal dysbiosis on frequent complications of advanced liver diseases can thusbe examined experimentally (cf. Figure 1).
Figure 1) experimental setupfor aim 2). Fecal microbiometransfer (FMT) from healthycontrols, infected animals,ACLD and ACLF animals,respectively, into healthyrecipients. The impact ofdysbiotic FMT on inflammationand liver disease progressionwill then be tested in FMTrecipients in injury models.GFM = germ-free mouse. LPS= lipopolysaccharide. DSS =dextran sodium sulfate.
To this end, we will test the hypothesis that ACLD-associated dysbiosis is an independently causaland therapeutically modifiable agent in the pathogenesis of ACLD-associated immune dys-function.Animals with an intestinal xenobiota are first generated by replacing the microbiota of healthyrecipient animals via FMT from Tak1Δhep animals - and vice versa as, a proof of concept.Subsequently, using models of acute colitis (induced by dextran sodium sulfate), septic shock(induced by lipopolysaccharide i.v.) or bacterial peritonitis (induced by E.coli i.p., analogous tospontaneous bacterial peritonitis in humans), we will assess the impact of FMT of eu- or dysbioticmicrobiota, respectively, on the host response to the corresponding immunological stimuli. Finally,we will examine the effects of the infection-associated pathobiota on the progression of liver diseasein the CCl4 model of inflammation-associated liver fibrosis, since acute extrahepatic inflammatorycomplications may promote the progression of ACLD and further worsen patients´ prognosis (3).
(3) The intestinal barrier as interface and target of intervention of the disrupted hostpathobiome interaction
In this part of the application, the focus will be on the (dys-)functionality of the intestinal barrier,mediating multi-layered interactions between host and microbiota, in the context of luminal dysbiosis.For this purpose, investigations on intestinal permeability, production of barrier-promoting factors(mucus, antimicrobial peptides), cell death processes in the epithelial layer and the release ofdamage-associated molecular patterns will be carried out in animals from parts 1A, 2A and 2B. Inaddition, we are planning FACS analyses of the intestinal mucosa at rest and under infectionconditions, with a focus on innate and adaptive immune cells, as well as sequencing analyzes of theintestinal mucosa and interactome analyzes of microbiota and intestinal immune cells in an unbiasedapproach. The aim of the investigations is to identify cellular effectors and signal cascades via whichthe pathobiota mediates its cross-border effects in liver diseases. Depending on the outcome of theinvestigations, targeted interventions that modulate immune cell recruitment (e.g. through integrinantagonists) or aberrant signaling of the pathobiota (e.g. Janus kinase inhibitors) are to beinvestigated in subsequent steps, and with regard to their suitability as therapeutic strategies incorresponding situations.
Project-related publications:
1. Albillos A, de Gottardi A, Rescigno M. The gut-liver axis in liver disease: Pathophysiological basis for therapy. J Hepatol. 2020Mar;72(3):558-577.
2. Bettermann K, Vucur M, Haybaeck J, et al. TAK1 suppresses a NEMO-dependent but NF-kappaB-independent pathway toliver cancer. Cancer Cell. 2010 May 18;17(5):481-496.
3. Engelmann C, Claria J, Szabo G, et al. Pathophysiology of decompensated cirrhosis: Portal hypertension, circulatorydysfunction, inflammation, metabolism and mitochondrial dysfunction. J Hepatol. 2021 Jul;75 Suppl 1:S49-S66.
-
Till Omansen (6. CS)
Lassa fever Pathogenesis
Project leader: Prof. Dr. med. univ. Michael Ramharter
Affiliation: Department of Tropical Medicine, Bernhard Nocht Institute for Tropical Medicine & I. Dep. of Medicine, University Medical Center Hamburg-Eppendorf
Background and preliminary data:
Lassa virus (LSAV) is an enveloped, negative-strand RNA virus causing Lassa fever (LF), a viral hemorrhagic fever (VHF). The disease is occurring in different regions in West Africa, with Nigeria being the hotspot. Hospitalized patients most commonly exhibit a sepsis-like disease phenotype with high inflammatory parameters, multi-organ failure and ultimately cardiovascular shock or a secondary neurological phenotype with seizures, meningitis and encephalopathy while kidney and liver function tests remain only moderately altered. To date it is unknown how LASV causes pathology in kidney, other organs and the central nervous system (CNS).
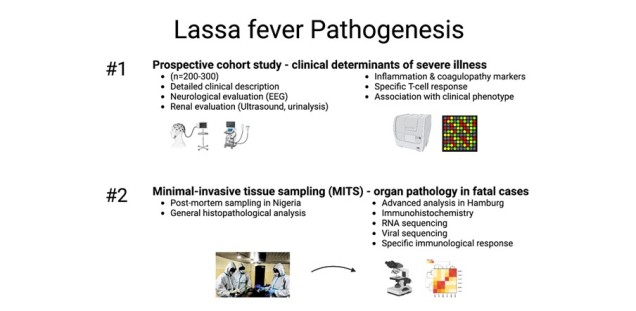
Within this research project we aim to decipher the pathogenesis of LF in a cohort of acutely infected patients in Nigeria and to test what the driving factors in the pathology of LF is. To this end, we recruit a prospective, observational cohort (n=300) of RT-PCR confirmed LF cases at our partner hospital in Nigeria. In addition, patients exhibiting neurologic or renal complications will be studied in detail with further non-invasive diagnostic tools such as EEG and ultrasound. The analysis for hypothesis testing will include measurement of inflammatory and hemostaseological biomarkers and analysis of the specific T-cell response.
This will be supplemented by a postmortem study on specimens collected with minimal invasive tissue sampling (MITS). MITS is a technique where tissue is collected using ultrasound-guided biopsies on the deceased. It offers the possibility to recover postmortem tissue for histopathological and other studies as conventional autopsy is not possible due to the biohazard in the case of a risk group 4 pathogen. In addition, MITS is culturally easier acceptable.
The study is carried out in close collaboration with the Department of Virology at the BNITM (further laboratory diagnostics and biosafety level 4 laboratory) and the Institute for Legal Medicine at UKE (MITS).
Hypothesis:
Severe & fatal Lassa fever is caused by specific dysregulation of the inflammatory response and coagulopathy
Aims and Work Programme:
In Aim #1, we will recruit a prospective, observational cohort of Lassa fever patients at our partner hospital in Edo State, Nigeria during outbreaks (n=200-300). Besides standard laboratory parameters and clinical examinations, blood samples will be taken every two days for the period of the hospitalization for further testing including of inflammatory and hemostasis markers. In addition, PBMCs are prepared on-site for further analysis of the specific T-cell response. Laboratory findings will be correlated to the disease severity and clinical phenotype to test which host response is associated with sever (and fatal) LF.
In Aim #2, postmortem tissue from all relevant organs and bodily fluids will be collected in a specialized facility within the isolation ward in Nigeria using the MITS protocol, which has recently been established on-site. Standard histopathological analysis is carried out in Nigeria while both fixated and native tissue is shipped to Hamburg for further analysis including immunohistochemistry, electron microscopy, and (spatial) transcriptomics.
Project-related publications:
Erameh et al. Prospective observational study on the pharmacokinetic properties of the Irrua ribavirin regimen used in routine clinical practice in patients with Lassa fever in Nigeria. BMJ Open . 2020 Apr 16;10(4):e036936. doi: 10.1136/bmjopen-2020-036936.
Groger et al. Pharmacokinetics of ribavirin in the treatment of Lassa fever: An observational clinical study at the Irrua Specialist Teaching Hospital, Edo State, Nigeria. Clin Infect Dis . 2022 Jul 26;ciac578. doi: 10.1093/cid/ciac578.
Strampe et al. Factors associated with progression to death in patients with Lassa fever in Nigeria: an observational study. Lancet Infect Dis . 2021 Jun;21(6):876-886. doi: 10.1016/S1473-3099(20)30737-4.
Akpede et al. Caseload and Case Fatality of Lassa Fever in Nigeria, 2001-2018: A Specialist Center's Experience and Its Implications. Front Public Health . 2019 Jun 25;7:170. doi: 10.3389/fpubh.2019.00170.
Eberhardt et al. Ribavirin for the treatment of Lassa fever: A systematic review and meta-analysis. Int J Infect Dis . 2019 Oct;87:15-20. doi: 10.1016/j.ijid.2019.07.015.
-
Fabian Heinrich (7. CS)
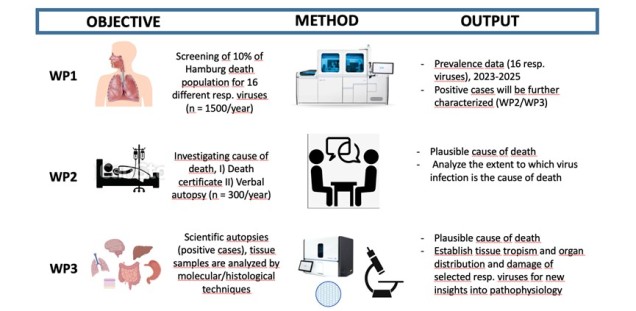
Analysis of the prevalence of respiratory viruses in deceased and investigation of a possible association with the cause of death - a representative cross-sectional study in the Hamburg area
Project leaders: Prof. Dr. Benjamin Ondruschka, Institute of Legal Medicine, UKE; Dr. Marc Lütgehetmann, Institute of Medical Microbiology, Virology and Hygiene, UKE.
Background and preliminary data: Respiratory tract infections (RTIs) contribute significantly to the global burden of infectious diseases. Key to understanding the morbidity and mortality of RTIs is also the involvement of organs beyond the respiratory tract, induced e.g., by systemic spread of the pathogens and associated with induction of severe inflammatory or septic processes (1). Robust data on the overall contribution of respiratory pathogens to mortality in the general population in Germany are scarce and probably underestimated because many deaths in residential and care settings are not investigated in this regard. A wide range of viruses can infect cells of the upper and/or lower respiratory tract, including epithelial-, endothelial-, tissue- and immune cells. Viral tropism depends on the expression of different receptors and co-receptors. E.g., for SARS-CoV-2, the receptor ACE2 and the co-factor TMPRTSS2 are essential, but several co-receptors, including vimentin, further promote viral entry. Interestingly, vimentin is upregulated in tissues in response to cell death and inflammation, further enhancing SARS-CoV-2 infection (1). Therefore, data on the tissue distribution of respiratory viruses in humans is important for understanding their pathophysiology, but currently knowledge in humans beyond influenza A and SARS-CoV-2 is very limited.
During the SARS-CoV-2 pandemic, we established high-throughput molecular workflows for the detection of respiratory viruses and bacteria in respiratory materials such as nasopharyngeal swabs, and bronchoalveolar lavage (2), allowing sensitive and cost effective screening of large cohorts. During the pandemic, we used these methods to investigate all cadavers (nasopharyngeal swabs) at the Institute of Legal Medicine Hamburg for the presence of SARS-CoV-2 RNA (1*). Positive cases were further analyzed through scientific autopsies and extensive tissue examination using molecular and histological methods. This approach has been shown to provide important and novel insights into the pathophysiology of this disease (2*-3*). The projects based on SARS-CoV-2 were funded by the German Research Network (NUM) within the projects DEFEAT PANDEMIcs and NATON. Current NUM funding covers exclusively infrastructure to enable future preparedness.
In order to go beyond SARS-CoV-2 epidemiology, we have screened (from 10/2021 to 12/2022) for 16 relevant respiratory viruses in a population-based approach, amounting to 35% of the death population in Hamburg (n = 7533 cases). Surprisingly, the positivity rate of samples with at least one respiratory virus detected was 21%. SARS-CoV-2 was still the dominant virus (12%), followed by rhino-/enterovirus (2%) and others with less than 1% (data unpublished). However, the extent to which virus detection represents a clinically relevant infection is an open question, and additional information on the cause of death - either anamnestic or biological - is needed to allow further differentiation. Based on the data and results of the previous work, we propose the following study.
Hypothesis: The number of respiratory infections as a cause of death is greatly underestimated in Germany.
Project and Goals: We will generate prevalence data of respiratory viruses in a representative cross-section of deceased people in Hamburg, evaluate virus-associated RTIs as a possible cause of death in this population, and analyze pathophysiological features, e.g., the tissue tropism and damage of the different viruses. This will re-evaluate the relevance of RTIs as cause of death and provide new insights into the pathophysiology of virus induced RTIs. We will address these objectives in three different work packages (WP).
- WP1: Estimation of the prevalence rates of different respiratory viruses in a population-based cross-sectional study of the deceased population in the metropolitan area of Hamburg (2023-2025).
- WP2: Determination of cause of death using death certificate documentation and, in positive cases, verbal autopsy.
- WP3: Determination of viral tissue tropism and damage as a potential cause of death by minimally invasive- and/or conventional autopsy in a subset of cases.
In WP1, we will use nasopharyngeal swabs (Universal transport medium, Copan) to screen corpses in the Hamburg area (Hamburg Crematoria and Institute for Legal Medicine, Hamburg, Germany). Based on the 2021/2022 data, we plan to screen at least 10% of the Hamburg deceased population (n = 1500 cases/year) starting in the 2023/24 winter season. Samples will be analyzed using established protocols for multiplex qPCR on highly automated systems (Cobas 6800/8800, Roche), allowing screening for 16 different respiratory viruses (ADV, Bocavirus, Influenza A/B, RSV, hMPV, PIV1-4, Rhino/Enterovirus, hCOV (229E, OC43, NL63, HKU1), SARS-CoV-2 using 3 qPCR reactions). Pooling protocols as described (4*) will be used to reduce material and financial costs.
In WP2, we plan to determine the plausible cause of death in all cases by 1) analyzing death certificate documentation and 2) additionally, in cases with positive virus detection (n= approx. 300/year), conducting a verbal autopsy (VA) with relatives of the deceased. The VA is a standardised WHO structured questionnaire used to determine and attribute causes of death. The process could be guided by the SmartVA software tool, which takes VA interview data as input and produces estimates of causes of death at the individual and population level (2).
In WP3, we plan to perform scientific autopsies in selected cases with detection of relevant amounts of virus NA (approximately 5-10 cases/for Influenza A_H1N1, Influenza A_H3N2, RSV, PIV1-4, hCOVs, Rhino/Enterovirus, ADV) after consent of the relatives using minimally invasive and/or conventional autopsy (as described in 5*). In addition to a rigorous review of the cause of death, a comprehensive sampling of various organs will be performed (pharynx, bronchus, lung, heart, liver, kidney, brain, colon, blood, vessels). Multiple samples per site will be collected (stored in liquid nitrogen for future projects by other scientists) and the tissue sample/organ will be analyzed by standard histology and by quantitative qPCR or digital PCR for virus RNA/DNA detection as previously described (2*-3*). The distribution of viral RNA/DNA in the tissues will be investigated by RNA in situ hybridisation (RNAscope) and viral protein by immunohistochemical staining (as described in 2*-3*).
Project related publications:
1. Heinrich F, Huter T, Mertens S, Lange P, Vering J, Heinemann A, Nörz DS, Hoffmann A, Aepfelbacher M, Ondruschka B*, Krasemann S*, Lütgehetmann M*. New Postmortem Perspective on Emerging SARS-CoV-2 Variants of Concern, Germany. Emerg Infect Dis. 2023;29(3):652-656.
2. Puelles VG*, Lütgehetmann M*, Lindenmeyer MT*, Sperhake JP*, Wong MN, Allweiss L, Chilla S, Heinemann A, Wanner N, Liu S, Braun F, Lu S, Pfefferle S, Schröder AS, Edler C, Gross O, Glatzel M, Wichmann D, Wiech T, Kluge S, Püschel K, Aepfelbacher M, Huber TB. Multiorgan and Renal Tropism of SARS-CoV-2. N Engl J Med. 2020;383(6):590-592.
3. Braun F*, Lütgehetmann M*, Pfefferle S*, Wong MN, Carsten A, Lindenmeyer MT, Nörz D, Heinrich F, Meißner K, Wichmann D, Kluge S, Gross O, Püschel K, Schröder AS, Edler C, Aepfelbacher M, Puelles VG, Huber TB. SARS-CoV-2 renal tropism associates with acute kidney injury. Lancet. 2020;396(10251):597-598.
4. Olearo F*, Nörz D*, Hoffman A, Grunwald M, Gatzemeyer K, Christner M, Both A, Campos CEB, Braun P, Andersen G, Pfefferle S, Zapf A, Aepfelbacher M, Knobloch JKM, Lütgehetmann M. Clinical performance and accuracy of a qPCR-based SARS-CoV-2 mass-screening workflow for healthcare-worker surveillance using pooled self-sampled gargling solutions: A cross-sectional study. J Infect. 2021;83(5):589-593.
5. Fitzek A, Schädler J, Dietz E, Ron A, Gerling M, Kammal AL, Lohner L, Falck C, Möbius D, Goebels H, Gerberding AL, Schröder AS, Sperhake JP, Klein A, Fröb D, Mushumba H, Wilmes S, Anders S, Kniep I, Heinrich F, Langenwalder F, Meißner K, Lange P, Zapf A, Püschel K, Heinemann A, Glatzel M, Matschke J, Aepfelbacher M, Lütgehetmann M, Steurer S, Thorns C, Edler C, Ondruschka B. Prospective postmortem evaluation of 735 consecutive SARS-CoV-2-associated death cases. Sci Rep. 2021;11(1):19342.
References:
1. Eslami N, Aghbash PS, Shamekh A, Entezari-Maleki T, Nahand JS, Sales AJ, Baghi HB. SARS-CoV-2: Receptor and co-receptor tropism probability. Curr Microbiol. 2022;79(5):133.
2. Pfefferle S, Reucher S, Nörz D, Lütgehetmann M. Evaluation of a quantitative RT-PCR assay for the detection of the emerging coronavirus SARS-CoV-2 using a high throughput system. Euro Surveill. 2020;25(9):2000152.
3. Hart JD, de André PA, de André CDS, Adair T, Barroso LP, Valongueiro S, et al. Validation of SmartVA using conventional autopsy: A study of adult deaths in Brazil. Lancet Reg. Health-Am. 2021;5:100081.
-
Lennart Hermanussen (8. CS)
Mechanisms underlying sex differences in HIV reservoir composition andsize in people living with HIV-1 on antiretroviral therapy
Project leader: Prof. Dr. Marcus Altfeld
Affiliation: Leibniz Institute of Virology, Hamburg
Background and preliminary data: Manifestations of HIV-1 infection differ between females and males. Previous studies haveshown that cis-gender women living with HIV-1 (WLWH) control viral replication better thancis-gender men living with HIV-1 (MLWH) in acute HIV-1 infection (Meditz et al., JID 2011). Incontrast, WLWH experience faster loss of CD4+ T cells and faster progression to AIDS duringuntreated chronic HIV-1 infection after controlling for the level of viral replication (Sterling etal., NEJM 2001). Increasing data indicate that these sex differences in the manifestations ofHIV-1 disease are mediated by sex-specific differences in antiviral immunity.
Please follow the link to download the project proposal.
-
Jan Peter Sutter (9. CS)
Analysis of the influence of gastrointestinal infections on host behavior via alteration of the gut microbiota, immune system and bacterial metabolites
Project leader: Prof. Dr. Samuel Huber and Prof. Dr. Matthias Kneussel
Affiliation: I. Department of Medicine, University Medical Center Hamburg-Eppendorf and Institute of Molecular Genetics, ZMNH
Background and preliminary data:
The gut-brain axis is a bidirectional communication network between the gut and the brain. It involves the gut microbiota, enteric nervous system, and immune system, influencing brain function including sociability, anxiety, and cognition. It has been shown that changes in the microbiota modulate host behaviour such as anxiety-related responses, social behaviour, and cognition. However, little is known about the influence of gastrointestinal infections on the infected individuals’ behaviour. Interestingly, one publication has postulated that infection with Citrobacter rodentium can cause anxiety-like symptoms that are probably mediated via vagal sensory neurons. However, a more recent study has shown that chronic inflammatory infections with Trichus muris induce anxiety-like behaviour in mice, which was present even after vagotomy. Furthermore, recent studies show that the immune system corresponds directly with the brain and modulates behaviour, and suggest that this might be mediated by altered cytokine production. For instance, maternal immune activation as a rodent model for autism induced a systemic release of IL-17a by Th17 cells in pregnant dams, resulting in impaired cortical patch development and behavioural abnormalities in the offspring. These effects were significantly increased in dams containing segmented filamentous bacteria which are known to potently induce Th17 cells in the intestine. Indeed, the phenotype could be reversed upon injection of IL-17A blocking antibodies. We have been studying the impact of the intestinal microbiota on the immune system and inflammatory diseases for years. Thus, as a first step and proof of concept, we found that different SPF microbiota are associated with a different intestinal immune cell composition (e.g. Th17 cell frequencies) and behaviour (e.g. anxiety) (unpublished data). Overall, those data suggest a crucial role of the immune system in mediating behavioural changes in the context of gastrointestinal infections. Whether this is actually the case and what mechanism(s) underlie this is, however, unclear and will be investigated in this project.
Hypothesis:
We hypothesize that gastrointestinal infections alter mice behaviour via modulating the intestinal microbiota and the immune system. Specifically, we hypothesize that infections promote anxiety, reduce sociability and cognition via microbiota alterations.
Our aim is to test this hypothesis and to decipher the underlying mechanism(s) in order to build the basis for targeted interventions for mental health conditions. To test the above-mentioned hypothesis, we aim to pursue the following objectives:
Aims and Work Programme:
- Analysis of behaviour including sociability, anxiety, and cognition of mice after infection with C. rodentium (model of bacterial gastrointestinal infection), and murine Cytomegalovirus (MCMV, viral model)
- Deciphering the functional role of the microbiota and bacterial metabolites on the immune system and host’s behaviour
Aim 1A: we will administer C. rodentium (a mouse model for E. coli infections) orally to 8 weeks old C57BL/6 wild-type mice. After 6 days (peak of disease) and 23 days (clearance of the pathogen) we will perform behavioural experiments including anxiety-like behaviour, sociability, and the cognition tests including a control group.
First, anxiety-like behaviour will be tested in an elevated-plus-maze with four arms, two closed and two open. A camera is used to measure the time spent in the open arms, indicating less anxious behaviour. Second, differences between the two groups in sociability are quantified in a social recognition test. After habituation in an empty three-chamber apparatus, mice are observed with an unknown intruder under one pencil cup while the other pencil cup is empty. The subject then encounters the now known intruder and a second unknown intruder. The number of entrances and the time spent in the different chambers, as well as the time spent sniffing each pencil cup, is recorded. Mice, which are normally social, prefer to spend time with conspecifics and are naturally more interested in novel objects or intruders, indicating social novelty and cognitive function. Deficits are seen, for example, in autism-like behaviour. Third, to further assess possible differences in cognition tasks, we perform an object recognition test. After several behavioural tests, the mice get used to the tests and tend to be less anxious in new situations, which is an advantage in cognitive tasks. The mice are placed in an open arena on the first day for habituation. On the training day, two similar objects are placed in different corners of the arena and the mouse is carefully placed in the middle to explore the arena. On the test day, one object is exchanged for a new, different object and the mouse explores the arena again. A camera is used to record the number of contacts and the time. Rodents tend to explore novel objects and this experiment is used to assess long-term recognition memory. Finally, we will also perform this experiment using a viral intestinal infection using MCMV to determine possible differences due to bacterial vs. viral gastrointestinal infections.
Aim 1B: we aim to decipher the role of the immune system in mediating the changes in behaviour upon a gastrointestinal infection. Specifically, we hypothesize that gastrointestinal infections can either directly or indirectly (via changes in the microbiota) promote the systemic release of proinflammatory cytokines and/or metabolites which are responsible for behavioural changes. To test this hypothesis, we will collect in the experiments of aim 1A intraepithelial lymphocytes and mesenteric lymph nodes, meninges, CNS, and blood. We will analyze the immune cell repertoire using flow cytometry (FACS) and defined cytokines as well as inflammatory markers (such as LPS) by using Enzyme-linked Immunosorbent Assay (ELISA). After separation of immune cells from tissues using Percoll gradient, quantitative determination of cell number is performed using automated cell counters (BioRad). This is followed by flow cytometry (FACS Fortessa, allows simultaneous measurement of 18 parameters) with extra-and intracellular staining for markers and cytokines (including for instance CD3, CD4, Foxp3, IL-10, IL-17A, IFN-γ, IL-4, IL-22, F4/80, CD11c, CD11b, CD14, Ly6G, Ly6C). The methods are already established in the laboratory. Furthermore, we will analyze the intestinal microbiota composition using metagenome sequencing. This analysis will be complemented by the metabolome analysis using biocrates.
Aim 2: we will use germ-free mice, which will be engrafted with the intestinal microbiota of the mice in aim 1 upon clearance of infection (A) and of patients upon clearance of a gastrointestinal infection (e.g. Campylobacter) (B). We will further study behaviour, immune cell composition, and metabolites essentially as described in aim 1B.
Long-term aim: to decipher the specific impact of metabolites, immune cells and cytokines on the observed effects to pave the way for targeted therapies.
Project-related publications (*own publications):
Bercik P et al (2010) Chronic gastrointestinal inflammation induces anxiety-like behavior and alters central nervous system biochemistry in mice. Gastroenterology 139(6):2102–2112 e1
Lyte M et al (2006) Induction of anxiety-like behavior in mice during the initial stages of infection with the agent of murine colonic hyperplasia Citrobacter rodentium. Physiol Behav 89(3):350–357
Lammert C et al (2018) Cutting Edge: Critical Roles for Microbiota-Mediated Regulation of the Immune System in a Prenatal Immune Activation Model of Autism. The Journal of Immunology 201 (3):845–850
Reed M et al. (2020) IL-17a promotes sociability in mouse models of neurodevelopmental disorders. Nature 577 (7789):249–253
*Tintelnot et al (2023) Microbiota-derived 3-IAA influences chemotherapy efficacy in pancreatic cancer. Nature 615(7950):168-174
-
Alina Ritter (10. CS)
Influence of Helicobacter pylori-induced alteration on the intestinal microbiota and on GEP-NET tumour growth
Project leader: Dr. Anna Nießen and Prof. Dr. Samuel Huber
Affiliation: Department of General, Visceral und Thoracic Surgery and I. Department of Medicine
Background and preliminary data:
Gastroenteropancreatic neuroendocrine tumours (GEP-NETs) are rare neoplasms that are thought to derive from hormone producing cells within the digestive tract. Due to their cellular origin, approximately 20% of GEP-NETs possess the ability to secrete hormones systemically and are thus characterised as functional NETs. While well-differentiated NETs display slow growth patterns as opposed to poorly differentiated NETs and Neuroendocrine Carcinomas (NECs), lymphatic and distant metastases can be present even in small, well-differentiated GEP-NETs. Prognosis of GEP-NET patients is highly dependent on the tumour stage and grade at primary diagnosis.
Helicobacter pylori (H. pylori) is a known risk factor for the development of gastric and duodenal NETs and H. pylori DNA has been found in pancreatic NET tumour tissue (1). It has been shown that H. pylori promotes colorectal carcinogenesis by altering the intestinal microbiota and inducing the oncogenic STAT signalling pathway – a pathway that likewise plays a role in GEP-NET progression (2, 3). Interestingly, GEP-NET patients display a distinct intestinal and intra-tumoral microbiota. Also, the composition of the intestinal microbiota plays a significant role in the treatment response of gastroenteropancreatic carcinoma (4). These data point towards a possible role of H. pylori and the intestinal microbiota in NET pathogenesis. However, little is known about the mechanisms by which H. pylori promotes NET development. Also, the influence on the intestinal microbiota and thus GEP-NET tumour growth is unknown.
Hypothesis:
The intestinal microbiota plays a key role in GEP-NET tumour development and growth. Infection with Helicobacter pylori alters the intestinal microbiota and can thereby enhance GEP-NET tumour growth.
Aims and Work Program:
- To determine the effect of H. pylori infection on the intestinal microbiota in GEP-NET patients.
- To determine the effect of H. pylori infection GEP-NET tumour growth and intestinal microbiota in vivo using a mouse model.
In Aim #1, we will analyse stool samples of GEP-NET patients to gain further insight into the composition of the intestinal microbiota and bacterial metabolites in these patients. Shotgun metagenomic sequencing as well as metabolomics will be applied on these stool samples (4, 5). Additionally, the past and present H. pylori infection status as well as possible H. pylori treatment and its influence on gut microbiota composition will be assessed in GEP-NET patients. Through analysis of tumour samples, its effect on immune cell infiltration into the tumour will be determined via immunohistochemistry. The intra-tumoral microbiota will likewise be analysed. The results will be correlated with tumour progression and responses to therapy of GEP-NET patients.
In Aim #2, a mouse model of GEP-NETs will be applied to assess the effect of infection with H. pylori in vivo. The composition of intestinal microbiota and their metabolites will be compared between H. pylori infected and uninfected animals. Additionally, the effect of H. pylori eradication on tumour growth in previously infected mice will be determined. Faecal microbiota transplantation from GEP-NET patients into germ-free mice will be used to further examine the effect of microbiota in vivo (4, 5).
Project-related publications:
1. Nam K, Nam SY, Park JC, Cho YS, Choi HS, Jung K, et al. Factors associated with gastric and duodenal neuroendocrine tumors: A multicenter case-control study. Dig Liver Dis. 2024;56(9):1592-8.
2. Ralser A, Dietl A, Jarosch S, Engelsberger V, Wanisch A, Janssen KP, et al. Helicobacter pylori promotes colorectal carcinogenesis by deregulating intestinal immunity and inducing a mucus-degrading microbiota signature. Gut. 2023;72(7):1258-70.
3. Amin T, Viol F, Krause J, Fahl M, Eggers C, Awwad F, et al. Cancer-Associated Fibroblasts Induce Proliferation and Therapeutic Resistance to Everolimus in Neuroendocrine Tumors through STAT3 Activation. Neuroendocrinology. 2023;113(5):501-18.
4. Tintelnot J, Xu Y, Lesker TR, Schönlein M, Konczalla L, Giannou AD, et al. Microbiota-derived 3-IAA influences chemotherapy efficacy in pancreatic cancer. Nature. 2023;615(7950):168-74.
5. Bedke T, Stumme F, Tomczak M, Steglich B, Jia R, Bohmann S, et al. Protective function of sclerosing cholangitis on IBD. Gut. 2024;73(8):1292-301.
-
Franziska Stallbaum (11. CS)
Understanding the influence of the gut microbiota and its metabolites on the host's susceptibility towards intestinal infections
Project leader: Prof. Dr. Samuel Huber
Affiliation: I. Department of Medicine, University Clinic Hamburg-Eppendorf
Background and preliminary data:
The microbial communities living in symbiosis with the host contribute to its health and regulation of the immune system. In contrast, microbial dysbiosis can lead to the dysregulation of bodily functions and the development of disease. Patients with inflammatory bowel disease (IBD) show distinct changes in the microbiota, which are associated with intestinal inflammation and an increased susceptibility toward both viral and bacterial infections. Adherent invasive E-coli (AIEC) is a well-known pathobiont, affecting 21-69% of patients with Cohn’s Disease (CD), compared to 0-19% of the general population. Infection with AIEC is associated with the development of ileitis and the progression of CD [1].The diet is a key modulator of the intestinal microbiota. Nutritional proteins and their metabolites can strongly influence the intestinal barrier function, the development of IBD, and the susceptibility towards gastrointestinal infections. For instance, a Western diet was shown to induce dysbiosis which led to altered epithelial barrier function and promoted AIEC colonization [2]. However, strategies to implement protective microbiota and reduce the risk of AIEC infection have not been fully explored. In preliminary work, we observed that a long-term gluten-free diet (LT-GFD) creates a protective microbiota, ameliorating Dextran Sodium sulfite (DSS)-induced colitis. However, how this “anti-inflammatory” microbiota and its metabolites influence the immune response toward gastrointestinal infections remains unclear. Addressing this question and identifying the characteristics and mechanisms of microbiota-mediated protection against gastrointestinal infections is the main goal of this proposal.
Hypothesis:
We hypothesize that metabolites derived from the gut microbiota after LT-GFD improve intestinal barrier function and consequently hold the capacity to protect from gastrointestinal infections.
Aims and work programme:
Aim #1: LT-GFD-Microbiota was shown to ameliorate colonic inflammation. We aim to investigate the influence of LT-GFD microbiota and its metabolites on the susceptibility towards AIEC infection in mice. In the first step, we will challenge mice with and without LT-GFD-derived microbiota by oral gavage of AIEC strains. Next, the susceptibility toward intestinal inflammation will be analyzed by monitoring weight loss and mucosal inflammation via endoscopy and end-point histology. The tissue-associated AIEC burden will be assessed by determining the colony-forming units after in vitro culture of tissue homogenates. The microbiota from ileum and colon samples will be analyzed by shotgun metagenomic sequencing. The metabolome will be deciphered using liquid gas chromatography and mass spectrometry. The intestinal barrier function will be assessed in vivo using P 4-kDa fluorescein isothiocyanate-conjugated dextran. RT-qPCR, histology, and immunohistochemistry will be used to describe changes of the intestinal epithelium. In addition, the isolation of cells of the adaptive and innate immune system will be performed and the immunophenotype will be described using Flow cytometry. [3-5]
Aim #2: The I. Department of Medicine at the University Medical Center Hamburg-Eppendorf, established a clinical trial in which individuals with inflammatory bowel disease undergo LT-GFD. First, we aim to investigate the prevalence of AIEC colonization and respective ileitis in patients with Crohn’s disease eating a LT-GFD. In this cohort, intestinal biopsies and stool samples will be analyzed to confirm our findings from aim #1 on the influence of LT-GFD derived microbiota on susceptibility towards AIEC infection. Subsequently, we will use fecalmicrobiota samples from CD patients before and after the LT-GFD intervention to reconstitute germ-free mice. These patient-specific gnotobiotic mice will be gavaged with AIEC strains, and a Crohn´s like disease model will be induced by blocking IL-10.
Project-related Publications:
1. Palmela, C., C. Chevarin, Z. Xu, J. Torres, G. Sevrin, R. Hirten, N. Barnich, S.C. Ng, and J.F. Colombel, Adherent-invasive Escherichia coli in inflammatory bowel disease. Gut, 2018. 67(3): p. 574-587.
2. Martinez-Medina, M., J. Denizot, N. Dreux, F. Robin, E. Billard, R. Bonnet, A. Darfeuille-Michaud, and N. Barnich, Western diet induces dysbiosis with increased E coli in CEABAC10 mice, alters host barrier function favouring AIEC colonisation. Gut, 2014. 63(1): p. 116-24.
3. Pelczar, P., M. Witkowski, L.G. Perez, J. Kempski, A.G. Hammel, L. Brockmann, D. Kleinschmidt, S. Wende, C. Haueis, T. Bedke, M. Witkowski, S. Krasemann, S. Steurer, C.J. Booth, P. Busch, A. Konig, U. Rauch, D. Benten, J.R. Izbicki, T. Rosch, A.W. Lohse, T. Strowig, N. Gagliani, R.A. Flavell, and S. Huber, A pathogenic role for T cell-derived IL-22BP in inflammatory bowel disease. Science, 2016. 354(6310): p. 358-362.
4. Bartsch, P., C. Kilian, M. Hellmig, H.J. Paust, A. Borchers, A. Sivayoganathan, L. Enk, Y. Zhao, N. Shaikh, H. Buttner, M.N. Wong, V.G. Puelles, T. Wiech, R. Flavell, T.B. Huber, J.E. Turner, S. Bonn, S. Huber, N. Gagliani, H.W. Mittrucker, H. Rohde, U. Panzer, and C.F. Krebs, Th17 cell plasticity towards a T-bet-dependent Th1 phenotype is required for bacterial control in Staphylococcus aureus infection. PLoS Pathog, 2022. 18(4): p. e1010430.
5. Huber, S., N. Gagliani, L.A. Zenewicz, F.J. Huber, L. Bosurgi, B. Hu, M. Hedl, W. Zhang, W. O'Connor, Jr., A.J. Murphy, D.M. Valenzuela, G.D. Yancopoulos, C.J. Booth, J.H. Cho, W. Ouyang, C. Abraham, and R.A. Flavell, IL-22BP is regulated by the inflammasome and modulates tumorigenesis in the intestine. Nature, 2012. 491(7423): p. 259-63.
-
Jonas Wagner (12. CS)
Role of tissue-specific T cells in organ-specific immunity
Project Leader: Prof. Dr. Nicola Gagliani
Affiliation: Hamburg Center for Translational Immunnology, Department of General, Visceral and Thoracic Surgery & I. Department of Medicine
Abstract:
Pathogens frequently gain entry into our bodies by penetrating surface tissues and establishing colonies in localized areas. When the pathogen is not controlled, their systemic spread can be lethal. We have helped establish the concept that a localized cell-mediated immune repose is fundamental to guarantee an efficient immunity (Vesely et al. Cell, 2019). Among the cells involved in this tissue-specific immunity, tissue-resident memory CD4 T cells (Trm) play a fundamental role. As each organ is profoundly different from each other organ, it is logical to hypothesize that Trm cells must adapt to the organ of residence to properly survive and function. Indeed, there is emerging evidence for transcriptional, phenotypical, and functional differences between Trm cells across different organs (Szabo et al. Nat. Comm., 2019; Wong et al. Immunity, 2016; Christo et al. Nat. Imm., 2021). However, the degree of CD4+ Trm organ-specific adaptation and the cause of this, especially in humans, has only just begun to be elucidated. This will be explored as part of this project.
-
Sophie Pflüger (13. CS)
Parvovirus B19 evolution and pathogenesis
Project leader: Prof. Dr. Nicole Fischer/ Dr. Marc Lütgehetmann
Affiliation: University Medical Center Hamburg-Eppendorf, Institute for Medical Microbiology, Virology and Hygiene
Background and preliminary data:
The clinical presentation of a parvovirus B19 (B19V) infection can range from an asymptomatic condition to life-threatening diseases such as myocarditis, aplastic crisis, and hydrops fetalis in pregnant women. Degree and severity of the disease correlate strongly with the immune status and hematopoietic maturation. Interestingly, B19V infection has also been associated with different diseases, including rheumatoid arthritis in adolescents and adults as well as kidney disease (PMIDs: 9529776; 17040883; 31202393). Primary B19V infection is common in childhood and small epidemics have been reported every few years (PMIDs: 14762186; 34391051). In young adolescents, B19V seroprevalence is approximately 50%, increasing to 80-90% with increasing age. Interestingly, Israel recently described a marked increase in symptomatic B19V infections. Children aged 6-11 years and pregnant women showed the most significant increase in B19V infection, when comparing the incidence rate ratio from 2015 – 2023 (PMID: 38005937).
B19V is a small, non-enveloped, single-stranded DNA virus with a genome of approximately 5.5 kb that encodes six viral proteins. In contrast to the previously thought strict tropism of B19V for erythroid progenitor cells in the bone marrow, additional receptors and co-receptors of B19V and infection of non-erythroid cells and vascular endothelial cells mediated by antibody enhanced uptake (PMID: 24807719) have been reported . This may explain the wide range of clinical manifestations of B19V infection (PMID: 29923512).
B19V is divided into three genotypes which form one serotype. The worldwide distribution of the genotypes varies, with genotype 1 accounting for 60% of infections in Europe, and genotype 1 accounting for nearly 95% of all described infections in Africa and Asia. Different genotypes are not only prevalent in different age groups, but have also been associated with different clinical presentations. Although there is insufficient data on the disease association of the different genotypes, genotype 2 has been described more frequently with cardiac manifestations. Regardless of genotype, viral genomic DNA was detected in multiple tissue types after primary infection in both symptomatic and asymptomatic individuals. However, a high viral load of genotype 1 was only detected in erythroid precursor cells of the human bone marrow (PMID: 29923512).
Limitations in cell culture systems, infection models as well as technical difficulties in genome recovery and sequencing have led to the fact that knowledge about the biology of B19V is still very limited. Currently, genetic diversity and many crucial aspects of B19V pathogenesis such as tissue tropism, persistence, and tissue damage remain very poorly understood.
Consistent with the observation of the largest so far described epidemic of B19V infections in Israel (PMID: 38005937), we have observed a significant increase in acute symptomatic B19V infections at the UKE with > 30 cases from Oct 2023 – Mar 2024, compared < 5 cases/year during the last 5 years. Of note, due to no reporting obligation for B19V infection in Germany benchmark data of B19V infections across Germany are missing. We propose that the current surge in B19V infections provides a unique opportunity to apply advanced molecular analytical and bioinformatical methods, many of them established during the SARS-CoV-2 pandemic (1-5), to improve our understanding of the disease.We propose to study the evolution of B19V in the current epidemic in the German population.
Our aims are:
1. To study the current epidemic of the virus in Hamburg: In the 30 cases observed in the UKE, viral load, viral diversity and viral infectivity will be characterized using retained saliva and serum samples. This will allow first conclusions on genotypes, intra-host diversity and viral load of this current outbreak.
2. To study genotypes, intra-host diversity and tissue tropism: We will complement the data obtained in 1. with data obtained from the UKE autopsy sample collection, allowing us to study primarily the tissue tropism of B19V.
3. To study possible disease association in immunocompromised hosts: As a long-term perspective, the data collected in this project will be analyzed and deepened in collaboration with the DZIF (TTU infections of immunocompromised hosts) on an existing large patient cohort.
WP1 (Virus kinetics and evolution): Analysis will be conducted on B19V positive samples from patients with acute disease (defined as anti-B19V-IgM positive, PCR > 10^6 IU/ml). Samples will be taken from the current epidemic (n=30 samples already retained) and acute samples from the past 10 years (n=10 samples). Whole genome sequences will be obtained using Parvovirus capture panel next generation sequencing. We will determine intra-host and inter-host diversity and follow the evolution of the virus within the current epidemic. Further, we will address prolonged, persistent infections in patients who fail to clear B19V DNA or experience a slow decline in levels (n=5) at 6 and 12 months after primary infection. This will give us first ideas on intra-host genome variability and possible viral factors accounting for persistence.
WP2 (infectivity and humoral immunity): Saliva and serum samples from at least 10 patients will be analyzed by digital PCR to quantify and correlate B19V DNA levels with anti-B19V IgG and IgA levels. To test for infectivity, selected samples will be employed for virus isolation using in vitro cell culture models (e.g., commercially available UT7/Epo-ST1 cells and/or IPSC-derived erythroid progenitor cells). Successfully isolated viruses will be further used in virus neutralization tests (VNTs) to assess the specificity and neutralizing capacity of the detected humoral immune responses.
WP3 (Tissue Tropism and Host Immune Response): To analyze long-term tissue tropism, archived autopsy samples from patients will be screened by qPCR of DNA pools covering multiple organs (intestine, heart, liver, kidney and spleen, and serum). B19V-positive pools will be deconstructed by digital PCR to define individual B19V load. Samples with high B19V DNA loads will be further analyzed by immunofluorescence staining and DNA in situ hybridization (DNA-Scope) to visualize infected cells in the organs and by capture panel sequencing as described in WP2 to obtain whole genome sequences. Finally, we will perform spatial transcriptomics (10x Xenium technology in collaboration with E. Whyler and M. Landthaler, MDC Berlin) on selected samples to identify B19V infected cell types and to analyze the host response to B19V infection/persistence at the single cell level.
Long-term perspective: To study B19V infection or reactivation of the virus in immunocompromised patients and the possible association with disease after kidney transplantation, we will apply to the DZIF Transplant Cohort for access to the biosampleshttps://www.dzif.de/de/arbeitsgruppe/transplantationskohorte. The transplant cohort includes more than 800 kidney transplant patients, more than half of whom had an infectious event in the first year after infection. While this cohort has been well studied for BK polyomavirus reactivation and herpesvirus reactivation (PMID: 35855001), no data are available yet on B19V.
Own references related to the proposal:
1) Heinrich F, Romich C, Zimmermann T, Kniep I, Fitzek A, Steurer S, Glatzel M, Nörz D, Günther T, Czech-Sioli M, Fischer N, Grundhoff A, Lütgehetmann M, Ondruschka B. Dying of VOC-202012/01 - multimodal investigations in a death case of the SARS-CoV-2 variant. Int J Legal Med. 2022 Jan;136(1):193-202.
2) Pfefferle S, Günther T, Kobbe R, Czech-Sioli M, Nörz D, Santer R, Oh J, Kluge S, Oestereich L, Peldschus K, Indenbirken D, Huang J, Grundhoff A, Aepfelbacher M, Knobloch JK, Lütgehetmann M, Fischer N. SARS Coronavirus-2 variant tracing within the first Coronavirus Disease 19 clusters in northern Germany. Clin Microbiol Infect. 2021 Jan;27(1):130.e5-130.e8.
3) Czech-Sioli M, Günther T, Robitaille A, Roggenkamp H, Büttner H, Indenbirken D, Christner M, Lütgehetmann M, Knobloch J, Aepfelbacher M, Grundhoff A, Fischer N. Integration of Sequencing and Epidemiologic Data for Surveillance of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Infections in a Tertiary-Care Hospital. Clin Infect Dis. 2023 Feb 8;76(3):e263-e273.
4) Heyer A, Günther T, Robitaille A, Lütgehetmann M, Addo MM, Jarczak D, Kluge S, Aepfelbacher M, Schulze Zur Wiesch J, Fischer N, Grundhoff A. Remdesivir-induced emergence of SARS-CoV2 variants in patients with prolonged infection. Cell Rep Med. 2022 Sep 20;3(9):100735.
5) Puelles VG*, Lütgehetmann M*, Lindenmeyer MT*, Sperhake JP*, Wong MN, Allweiss L, Chilla S, Heinemann A, Wanner N, Liu S, Braun F, Lu S, Pfefferle S, Schröder AS, Edler C, Gross O, Glatzel M, Wichmann D, Wiech T, Kluge S, Pueschel K, Aepfelbacher M, Huber TB. Multiorgan and Renal Tropism of SARS-CoV-2. N Engl J Med. 2020 Aug 6;383(6):590-592.
-
Alexandra Linke (14.CS)
Immune regulation and impaired vaccine responsiveness in autoimmune liver diseases
Project leaders: Prof. Dr. Ansgar W. Lohse/ Dr. Johannes Hartl
Affiliation: I. Medizinische Klinik und Poliklinik, UKE
Background and preliminary data:
For patients with autoimmune disease vaccinations against common pathogens are generally recommended, especially when, or prior to, receiving immunosuppressive therapy. However, we do not really know, if patients with autoimmune diseases can mount a normal anti-vaccine response, what influence immunosuppressive therapy has on the anti-vaccine response, and how disease activity and vaccine responsiveness are inter-related(1).
For autoimmune liver diseases, in particular for autoimmune hepatitis (AIH), we have some data suggesting there is a relevant impairment of vaccine responsiveness, that this is present both in untreated patients and in patients undergoing immunosuppressive therapy, and that this is distinct from the other major autoimmune liver diseases, primary biliary cholangitis and primary sclerosing cholangitis (2, 3). In fact, it was shown already 30 years ago that patients with newly diagnosed untreated AIH show an almost completely abrogated response to a tetanus toxoid booster vaccination(4). Very recently we could show that response to SARS-CoV2 vaccine was impaired in AIH patients, both in the humoral and the cellular arm of the vaccine response, and that this impairment was present to a similar degree in patients receiving immunosuppressive therapy compared to untreated AIH patients in remission(2).
Hypothesis:
The hypothesis is, that an impairment of the anti-vaccine response is an expression of a generalized autoregulatory immune response of the patients in an attempt to dampen their autoimmune reaction against liver target antigens. It is furthermore hypothesized that this autoregulatory response is largely mediated by interleukin 10 and transforming growth factor beta, two key cytokine found to be elevated in AIH. Finally, it is hypothesized that the characteristic elevation of IgG in AIH is an expression of this autoregulatory response, which is therefore specific for AIH and not to be found in other liver diseases, autoimmune or metabolic.
Aims and work programme:
The aim of the suggested project is to assess this phenomenon of altered anti-vaccine responsiveness more closely looking at different vaccines (influenza, pneumococci, tetanus and SARS-CoV2 – all recommended vaccinations in AIH) deciphering the anti-vaccine response on a molecular level, and in particular to correlate the anti-vaccine response with clinical parameters such as activity of AIH, disease phase (are the patients on the ascending loop of AIH activity, or are they in a descending phase?), and correlate with key cytokine expression. A particular focus will be on IgG-levels, which are thought by some to be an expression of AIH activity(5), while more recent data suggest that raised IgG-levels are a consequence of the immunoregulatory response of the host, mediated largely by IL10 and TGFß, leading to increased production of IgG. The study will then put a particular focus on a group of patients with AIH, in whom immunosuppressive therapy has recently been tapered, as some of these will relapse, while others stay in remission, the twodisease stages being associated with distinct immune profiles, whose kinetics will be characterized and related to vaccine responsiveness. Control patients will be PSC, PBC and non-immune liver diseases (NASH).
Publications:
1. Pape S, Snijders R, Gevers TJG, Chazouilleres O, Dalekos GN, Hirschfield GM, et al. Systematic review of response criteria and endpoints in autoimmune hepatitis by the International Autoimmune Hepatitis Group. J Hepatol. 2022;76(4):841-9.
2. Duengelhoef P, Hartl J, Ruther D, Steinmann S, Brehm TT, Weltzsch JP, et al. SARS-CoV-2 vaccination response in patients with autoimmune hepatitis and autoimmune cholestatic liver disease. United European Gastroenterol J. 2022;10(3):319-29.
3. Hartl J, Ruther DF, Duengelhoef PM, Brehm TT, Steinmann S, Weltzsch JP, et al. Analysis of the humoral and cellular response after the third COVID-19 vaccination in patients with autoimmune hepatitis. Liver Int. 2023;43(2):393-400.
4. Lohse AW, Kogel M, Meyer zum Buschenfelde KH. Evidence for spontaneous immunosuppression in autoimmune hepatitis. Hepatology. 1995;22(2):381-8.
5. Hartl J, Miquel R, Zachou K, Wong GW, Asghar A, Pape S, et al. Features and outcome of AIH patients without elevation of IgG. JHEP Rep. 2020;2(3):100094.
-
Dimitra Zazara-Giannou (15. CS)
Respiratory tract infections during early childhood as a consequence of an obese intrauterine environment: Transgenerational effects on trained immunity
Project leader: Prof. Dr. Petra Arck
Affiliation: Department of Obstetrics and Fetal Medicine & Hamburg Center for Translational Immunology, University Medical Center Hamburg
Background and preliminary data:
The prevalence of overweight and obesity is increasing worldwide, which also affect pregnant women. To date, more than 30% of pregnant women who are overweight or obese. Maternal obesity has been associated with adverse consequences on children’s health. In this context, epidemiological studies highlight an increased risk for lower respiratory tract infections early in life in children born to obese mothers. Lower respiratory tract infections early in life have detrimental long-term consequences, as they confer a significantly higher risk for chronic respiratory diseases later in life. These can range from wheezing and asthma during childhood to chronic obstructive pulmonary disease in adult offspring. It has been hypothesized that these risks do not result from genetic or maternal lifestyle factors, but are triggered by the modulation of fetal development within the obese intrauterine environment. However, molecular mechanisms that underlie the increased risk for lower respiratory tract infections and subsequent respiratory diseases in offspring born to mothers with obesity are still largely unknown. Since respiratory tract infections are the world’s leading cause of death in young children and respiratory diseases a main reason for hospitalization, translational research approaches are urgently required to reduce such risks and develop prenatal primary prevention strategies.
Maternal obesity is associated with numerous changes, such as higher levels of leptin, high sensitivity C-reactive protein and chronic low-grade inflammation. These maternal markers, can pass the placental barrier and enter the foetal circulation. In the fetus, these markers can interfere with the development of fetal organs, such as the lung, and the fetal immune system.
In this context, the development of the innate immune system may be of particular relevance. As shown by our group and others, the innate immune system emerges from erythro-myeloid progenitors in the yolk sac, which subsequently seed into foetal organs, such as the lung, skin, brain, liver and other organs, where they ultimately differentiate into macrophages, e.g., alveolar macrophages in the lung. Here, alveolar macrophages maintain lung tissue homoeostasis and orchestrate immunity upon pathogen encounter after birth. Interestingly, alveolar macrophages are tissue-resident cells, they mainly self-renew and are only marginally replenished by haematopoietic stem cell-derived macrophages. Thus, an altered profile of alveolar macrophages - triggered by an obese intrauterine environment – can have consequences throughout life. Given that an obese intrauterine environment increases the risks for lower respiratory tract infections, asthma and COPD, as identified in epidemiological studies, investigating the development and function of alveolar macrophages is key to decode how maternal obesity alters children’s pathogen response in the lung. In this context, modulation of the chromatin state and subsequently skew of macrophages function, a phenomenon is known as innate immune memory, is of particular interest, as it is unknown if an obese intrauterine environment and associated low-grade inflammation can induce such epigenetic changes in fetal innate immune cells. Such causality, if confirmed, can the serve as an important target to reduce the risk for lower respiratory tract infections during early childhood.
Taken together, the present proposal will improve our understanding of the immune response to pathogens causing lower respiratory tract infections in the highly vulnerable population of small children. Our group has a broad experience in preclinical models, along with the access to human biosamples and metadata from mothers and children in our longitudinal pregnancy cohort PRINCE. From the latter, we recently provided evidence that fetal lung growth can be monitored sonographically and linked to early life infections. Our established mouse models include assessment of lung development and the induction and modulation of early life infections and asthma.
Hypothesis:
An obese intrauterine environment affects fetal lung and innate immune development, e.g., via epigenetic changes such as trained immunity, which has functional consequences for immunity against pathogens causing lower respiratory tract infections during early childhood.
Aims and work programme:
Aim 1: To investigate the interaction between maternal obesity and fetal lung and immune development in mice. Maternal obesity will be induced by high fat diet prior to mating, maternal obesity-triggered inflammation signature will be characterized. Fetal lung growth and differentiation of monocyte precursor cells to alveolar macrophages during fetal development will be evaluated (lung morphometry, flow cytometry, chromatin state, infection with Streptococcus pneumoniae and influenza A virus, possibly followed by deep immune phenotyping of cells via scRNASeq). Based on these outcomes, experiments using e.g., transgenic mice lacking specific receptors on innate immune (precursor) cells can be designed. All methods are established in our team or our collaborators.
Aim 2: To assess the interaction between maternal obesity, immune development and infection risks in children using data and samples from the PRINCE study, amended by the establishment of clonal epithelial lung organoids derived from epithelial stem/progenitor cells isolated from amniotic fluid in humans[xvii].
Project-related publications:
Arck PC, Hecher K. Fetomaternal immune cross-talk and its consequences for maternal and offspring's health. Nat Med, 2013.
Kinder JM, Stelzer IA, Arck PC, Way SS. Immunological implications of pregnancy-induced microchimerism. Nat Rev Immunol, Aug;17(8):483-494, 2017.
Zazara DE, Wegmann M, Giannou AD, Hierweger AM, Alawi M, Thiele K, Huber S, Pincus M, Muntau AC, Solano ME, Arck PC. A prenatally disrupted airway epithelium orchestrates the fetal origin of asthma in mice. J Allergy Clin Immunol. 45(6):1641-1654, 2020.
Stelzer IA, Urbschat C, Schepanski S, Thiele K, Triviai I, Wieczorek A, Alawi M, Ohnezeit D, Kottlau J, Huang J, Fischer N, Mittrücker HW, Solano ME, Fehse B, Diemert A, Stahl FR, Arck PC. Vertically transferred maternal immune cells promote neonatal immunity against early life infections. Nat Commun, 12(1):4706, 2021.
Zazara DE, Giannou O, Schepanski S, Pagenkemper M, Giannou AD, Pincus M, Belios I, Bonn S, Muntau AC, Hecher K, Diemert A, Arck PC. Fetal lung growth predicts the risk for early-life respiratory infections and childhood asthma. World J Pediatr. doi: 10.1007/s12519-023-00782-y. Epub ahead of print, 2024.
-
Kevin Paul (16. CS)
Organ- and age-specific host defense responses in adenovirus-infections cells
Project leader: Prof. Dr. Madeleine J. Bunders
Affiliation: III. Department of Medicine, UKE
Background and preliminary data:
Human adenovirus (HAdV) infections are an important cause of respiratory, hepatic and gastrointestinal infections. Especially children and immune-compromised adults can suffer from severe adenovirus infection and reactivation. HAdV comprise of a large family of DNA viruses of which most express proteins that exhibit exceptional capacities to evade immune control of infected cells and establish latent infection, such as E1 and E3 proteins. Until now most studies investigating HAdV infection are performed in cell lines, however our recent findings, using human tissue-based organoid systems, demonstrate that cell lines poorly model HAdV infection in human cells and HAdV-mediated regulation of host defense responses. Using intestinal organoid systems we were able to identify a new target (HLA-F) identifying HAdV-infected epithelial cells to NK cells, which provided children following hematopoietic stem cell transplantion protection by KIR3DS1+ NK cells against severe HAdV reactivation. However, the HAdV strains infect multiple organs resulting in organ-specific infection and tissue damage. To be able to investigate organ-specific HAdV infection, we have extended the HAdV organoids models to study HAdV infection in airway, liver and intestinal organoid systems. Our preliminary studies of the different organ systems indicate that age and organ-specific dynamics of HAdV infection exist and HAdV proteins show organ-specific modulation of host defense mechanisms relating to specific populations at risk. In this project we will investigate organ-specific and age-specific HAdV infection to understand organ-specific HAdV disease.
Hypothesis:
HAdV mediates organ and age-specific disease in humans
Aims and Work Programme:
1. To assess the correlation between organ and age specificity of HAdV infectivity and viral production. 2. To identify organ-specific and shared pathways in epithelial cells affected by HAdV that mediate viral control in children and adults. In Aim #1, organoids from gut, lung and liver tissue are generated from infant and adult donors. Organoids will be infected with a specific serotypes including serotypes with specific tropism to an organ (e.g. HAdV40/41) to serotypes with a capacity to infect a broader range of organs (e.g. HAdV5). Infection will be quantified by flow cytometry and microscopy (viral proteins) and viral DNA in a time course. Cellular characteristics of HAdV-infected cells in organoid systems will be determined by flow cytometry and microscopy. Data will be analyzed using regression models to assess organ and age-specific risk factors for viral infectivity and production. To validate these in vitro studies risk factors for HAdV infection will next be tested using data from large cohorts of individuals undergoing HSCT correlating viral load and organ specific symptoms. Together, these tissue-based and cohort based studies will identify characteristics of patients and epithelial cells susceptible for specific-HAdV serotypes in human organs and HAdV dynamics during infection. In Aim #2, the generation of robust HAdV infection organoid systems now allows to further determine organ- and age-specific pathways affected by HAdV. To this end, scRNAseq analyses of organoids systems have been established and will be employed to 3 organ organoid systems generated from infants and adults with 3 different serotypes. Computational tools have been developed to perform transcriptomics analyses of HAdV-infected cells and compare to HAdV (RNA) negative cells from the same organoid culture. Upon identification of specific pathways these will be validated using blocking antibodies or gene-edited organoids using Crispr/Cas9. Together these studies will identify pathways shared between organs that are affected by specific HAdV strains as well as organ specific pathways that are associated with HAdV disease in children and adults.
Project-related publications: (max. 5) * shared
1. Möller KJ*, Wegner L*, Malsy J*, et int, Bunders MJ. Expanded ILC2s in human infant intestines promote tissue-growth. Mucosal Immunol. 2023; doi: 10.1016/j.mucimm.2023.04.004. Online ahead of print.
2. Jordan-Paiz A*, Martrus G*, Fenja Steinert*, et int, Bunders MJ. CXCR5+PD-1++ CD4+ T cells colonize infant intestines early in life and promote B cell maturation. Cell Mol Immunol. 2023; 20(2):201-213. doi: 10.1038/s41423-022-00944-4.
3. Jung JM, Ching W, et int, Altfeld M*, Belderbos M*, Dobner T*, Bunders MJ*. KIR3DS1 directs NK cell-mediated protection against human adenovirus infections. Sci Immunol. 2021; 6(63):eabe2942. doi: 10.1126/sciimmunol.abe2942.
4. Sagebiel AF, Steinert F, et int, Bunders MJ. Tissue-resident Eomes(+) NK cells are the major innate lymphoid cell population in human infant intestine. Nat Commun.2019; 10(1):975. doi: 10.1038/s41467-018-08267-7.
5. Schreurs RRCE, Baumdick ME*, Sagebiel AF*, et int, Bunders MJ. Human fetal TNF-α-cytokine-producing CD4(+) effector memory T cells promote intestinal development and mediate inflammation early in life. Immunity.2019; 50(2):462-476.e8. doi: 10.1016/j.immuni.2018.12.010.
-
Tamara Nordmann (17. CS)
Clinical development of oxfendazole for the treatment of Loa loa in central African Gabon
Project leader: Prof. Dr. med. univ. Michael Ramharter
Affiliation: Department of Clinical Research, Bernhard Nicht Institute for Tropical Medicine & I. Dep. of Medicine, Div of Tropical Medicine, University Medical Center Hamburg-Eppendorf
Background and preliminary data:
Loa loa – the African eye worm – is a highly neglected helminthic infectious disease prevalent in Central and parts of West Africa. Loiasis mainly affects rural populations in the rainforest and savannah regions where the principal vector Chrysops acts as efficient vector. Current anthelminthic drugs are insufficient options for the treatment and control of loiasis due to the necessity of prolonged treatment regimens or important safety concerns with life-threatening adverse drug reactions.
Oxfendazole is among the most promising novel anthelminthic drug candidates for the clinical development for the treatment of loiasis. Oxfendazole has been successfully used for decades in veterinary medicine and is currently under clinical development as repurposed drug for human use by Drugs for Neglected Diseases Initiative (DNDi). It has shown excellent pharmacodynamic activity against model filarial pathogens in preclinical animal models and has successfully finished a clinical phase I program.
In this project a series of clinical phase II and III trials funded by the German Center for Infection Research and the EU program EDCTP3/GH JU will be conducted in collaboration with the CERMEL in Lambaréné, Gabon, DNDi and the University of Bonn. The clinical trials will be conducted at the CERMEL and its satellite sites in remote rural Gabon.
Hypothesis:
The objective of the clinical phase II/III development program is to perform a dose finding clinical trial assessing the safety, tolerability and efficacy of oxfendazole for the treatment of loiasis. Based on the result of this clinical trial, a subsequent clinical phase II/III trial will be conducted as part of an international basket trial program to further evaluate the pharmacodynamics, pharmacokinetics, and safety/tolerability of oxfendazole treatment.
Aims and Work Programme:
- Randomized controlled dose finding clinical phase II trial for the treatment of loiasis
- Assessment of the safety/tolerability of oxfendazole therapy on loiasis
- Assessment of efficacy of oxfendazole therapy on L. loa microfilaraemia
- Assessment of oxfendazole therapy on concommittant infections with Mansonella spp. And soil transmitted helminths.
- Characterization of the pharmacokinetics of oxfendazole in fasted and fed states for single and multiple dose regimens
- Randomized controlled clinical phase II/III trial assessing the efficacy, safety, tolerability of an optimized oxfendazole regimen for the treatment of loiasis in an adaptive clinical trial design.
Candidates with the following background will be best suited for this clinical trial program
- Candidates willing to work for several months at the partner institution CERMEL, Gabon, as investigators in the conduct of the above mentioned clinical trials
- Previous experience in clinical drug trials and work in sub-Sahara Africa is welcome
- Basic (or more advanced) language skills in French are helpful for this project in francophone Gabon
- Previous experience in medical research in the field of human parasitology/tropical medicine is encouraged
Project-related publications:
Distinct loiasis infection states and associated clinical and hematological manifestations in patients from Gabon. Veletzky L, Eberhardt KA, Hergeth J, Stelzl DR, Zoleko Manego R, Mombo-Ngoma G, Kreuzmair R, Burger G, Adegnika AA, Agnandji ST, Matsiegui PB, Boussinesq M, Mordmüller B, Ramharter M. PLoS Negl Trop Dis. 2022 Sep 19;16(9):e0010793.
Performance of Field's Stain Compared with Conventional Giemsa Stain for the Rapid Detection of Blood Microfilariae in Gabon. Mbassi FE, Mombo-Ngoma G, Ndoumba WN, Yovo EK, Eberhardt KA, Mbassi DE, Adegnika AA, Agnandji ST, Bouyou-Akotet MK, Ramharter M, Zoleko-Manego R. Am J Trop Med Hyg. 2022 Jul 5;107(2):383-7.
Diagnostic performance of capillary and venous blood samples in the detection of Loa loa and Mansonella perstans microfilaraemia using light microscopy. Mischlinger J, Manego RZ, Mombo-Ngoma G, Ekoka Mbassi D, Hackbarth N, Ekoka Mbassi FA, Davi SD, Kreuzmair R, Veletzky L, Hergeth J, Ndoumba WN, Pitzinger P, Groger M, Matsiegui PB, Adegnika AA, Agnandji ST, Lell B, Ramharter M. PLoS Negl Trop Dis. 2021 Aug 16;15(8):e0009623.
Burden of disease in Gabon caused by loiasis: a cross-sectional survey. Veletzky L, Hergeth J, Stelzl DR, Mischlinger J, Manego RZ, Mombo-Ngoma G, McCall MBB, Adegnika AA, Agnandji ST, Metzger WG, Matsiegui PB, Lagler H, Mordmüller B, Budke C, Ramharter M. Lancet Infect Dis. 2020 Nov;20(11):1339-1346.
Behavioural and clinical predictors for Loiasis. Mischlinger J, Veletzky L, Tazemda-Kuitsouc GB, Pitzinger P, Matsegui PB, Gmeiner M, Lagler H, Gebru T, Held J, Mordmüller B, Ramharter M. J Glob Health. 2018 Jun;8(1):010413.
- Randomized controlled dose finding clinical phase II trial for the treatment of loiasis
-
Henrike Großhans (18. CS)
Neuron-intrinsic immune responses during viral encephalitis
Project leader: Prof. Dr. Manuel Friese
Affiliation: Institute for Neuroimmunology and Multiple Sclerosis
Background and preliminary data:
Current State of Research
Neurotropic viral infections of the central nervous system (CNS) present a significantworldwide health risk and a neglected medical problem. Viral encephalitis, mostly caused byherpes simplex virus (HSV), varicella–zoster virus, enteroviruses, or arboviruses, is ignitinginflammation of the CNS parenchyma with associated neurological dysfunction and highmortality (1). Previous studies on CNS-specific immunity to viruses have primarily focused onroles of circulating leukocytes and resident microglial cells. Traditionally, neurons were viewedas passive victims of viral infection, dependent on external antiviral mechanisms for protection.However, over the past decade, there has been a growing recognition that neurons also havecell-intrinsic mechanisms to combat viruses (2).
The CNS has multi-layered defenses against viral infections, but once breached,damage is rapid and severe due to limited neuronal regenerative capacity. Pattern recognitionreceptors (PRRs) initiate the immune response by detecting viral molecules, triggering thesynthesis of antiviral proteins and cytokines like type I (IFN-α/β) and II interferons (IFN-γ). Ifintrinsic defenses fail, sentinel cells are activated, producing more cytokines and triggeringadaptive immunity. An overreaction, however, can cause immunopathology which are similarto conditions occurring in autoimmune CNS diseases (3).
Host-pathogen co-evolution has driven immune gene diversification (4), but extendedinnate immune responses can damage organs like the CNS. Interferon-stimulated genes(ISGs) upregulate antiviral activities but can disrupt host cell homeostasis. Recently, neuronaltranscript profiling under sterile inflammatory conditions identified apolipoprotein L6 (APOL6),a member of the ISG family, as highly induced during inflammation in neurons (5). APOL6'sfunction is largely unknown, but ISG screenings identified APOL6 as an inhibitor of coxsackieB virus and poliovirus, members of the picornavirus family, in cell culture (6). Furthermore,several studies associate the antiviral properties of APOL6 with additional types of viruses,including SARS-CoV-2 and HIV-1. Thus, APOL6 is a paradigmatic example of a neuronintrinsicdefense mechanism against neurotropic viruses.
Preliminary Work
In neuronal cells, CRISPRa-induced APOL6 expression provided significant protection againstTheiler's murine encephalomyelitis virus (TMEV), comparable to IFN-γ pretreatment.
Moreover, subcellular localization and interactome studies showed APOL6 to belocalized and interact with proteins at mitochondria and the endoplasmatic reticulum (ER),indicating association with mitochondria-associated ER membranes (MAMs), crucial forantiviral responses. We found overexpression of APOL6 to affect cell viability, ER calciumlevels, and the mitochondrial membrane potential, suggesting interference in calciumhomeostasis and mitochondrial function, which is crucial for certain virus life cycles. CRISPRamediatedAPOL6 upregulation revealed the induction of antiviral pathways, indicating APOL6'srole in regulating interferon-signaling pathways.
To study APOL6 in vivo, we established Apol6-deficient mice and developed methodsfor neuron-specific overexpression of APOL6 using adeno-associated viruses. This approach,combined with our CRISPR technology, will allow us to further explore APOL6's role in CNSinfections and inflammation, but also to expand our toolbox to other neuron-intrinsic ISGs.
Hypothesis:
We hypothesize that neuron-intrinsic immune responses to neurotropic viruses are in adelicate balance between virus containment and neuronal survival and self-damage, and thatneurons have evolved specific defense mechanisms against viruses due to their uniqueproperties.
Aims and Work Programme:
Viral encephalitis, with HSV-1 being the most common causative agent, is a major neglectedmedical issue. Additionally, emerging viral infections present a high risk for developing newtypes of encephalopathies, highlighting the need for targeted therapeutic strategies. UsingAPOL6 as a prime example, we aim to enhance our understanding of the antiviral, neuronintrinsicimmune response and to identify novel innate immune genes that could be leveragedfor developing antiviral therapies for the CNS.
WP1: Deciphering the molecular mechanism of APOL6’s antiviral function.
This work package builds upon our laboratory's research on the antiviral and neurotoxicproperties of APOL6 within the SFB1648. We have already established a variety of methodsin this context, including gene knockout and activation in vitro and in vivo using CRISPR-Castechnology, neuron-specific delivery in vivo using recombinant AAVs, mouse primary neuronalcultures, and human induced pluripotent stem cell (hiPSC)-derived neuronal cultures, coupledwith longitudinal cell viability assays. These techniques provide an excellent foundation for theIDfellow to familiarize themselves with all relevant methodologies while simultaneously gainingnew insights into this neuron-intrinsic antiviral protein. In this work package, we will investigatethe molecular mechanisms by which APOL6 mediates its antiviral and neurotoxic activities.We will utilize mutational and CRISPR-KO screenings to identify the critical amino acids in theAPOL6 protein and its downstream effector genes.
WP2: Identification of neuron-intrinsic antiviral immune response genes.
This work package aims to extend our knowledge about neuron-intrinsic ISGs to additionalunknown candidates. Thus, here we want to identify novel factors involved in the neuronintrinsicantiviral immune response. We will use murine and human neuronal cell lines,employing knockout and activation strategies. (i) Utilizing an ISG CRISPR-KO library, we willperform gene knockouts in combination with interferon treatment to render the cells resistant,followed by viral infection. Genes whose loss results in susceptibility will be identified throughnext-generation sequencing. (ii) Employing a CRISPRa library, we will activate genes followedby viral infection. Genes whose activation leads to resistance will be identified through nextgenerationsequencing. The identified candidate genes from both strategies will then bevalidated in murine and hiPSC-derived neuronal cultures and can be developed by the IDfellowinto an independent mechanistic project.
Project-related publications:
1. A. Venkatesan, B. D. Michael, J. C. Probasco, R. G. Geocadin, T. Solomon, Acuteencephalitis in immunocompetent adults. The Lancet 393, 702–716 (2019).
2. S.-Y. Zhang, O. Harschnitz, L. Studer, J.-L. Casanova, Neuron-intrinsic immunity to virusesin mice and humans. Curr. Opin. Immunol. 72, 309–317 (2021).
3. M. S. Woo, et al., STING orchestrates the neuronal inflammatory stress response in multiplesclerosis. Cell 187, 4043-4060 (2024).
4. E. E. Smith, H. S. Malik, The apolipoprotein L family of programmed cell death and immunitygenes rapidly evolved in primates at discrete sites of host-pathogen interactions. GenomeRes. 19, 850–858 (2009).
5. B. Schattling, et al., Bassoon proteinopathy drives neurodegeneration in multiple sclerosis.Nat. Neurosci. 22, 887–896 (2019).
6. J. W. Schoggins, et al., Pan-viral specificity of IFN-induced genes reveals new roles forcGAS in innate immunity. Nature 505, 691–695 (2014).
-
Matin Kohsar (19. CS)
Deciphering coagulation disorder in Lassa fever
Project lead: Dr. Till Omansen
Affiliation: Center for Tropical Medicine, Bernhard Nocht Institute for Tropical Medicine & I. Dep. of Medicine, University Medical Center Hamburg-Eppendorf; Department of Virology, Bernhard Nocht Institute for Tropical Medicine
Background and preliminary data:
Lassa fever is a viral hemorrhagic fever endemic in West Africa. Case fatality rates in hospitalized patients are high, ranging from 10 to 30%. As a viral hemorrhagic fever, bleeding complications are a major feature of severe Lassa fever - albeit seemingly not as severe as in Ebola virus disease - and associated with high mortality. Still, little is known about the actual frequency of coagulation disorder in Lassa fever, its clinical relevance in patient management, and its pathogenesis.
Experimental data from non-human primates highlighted the role of impaired hepatic synthesis of coagulations factors, circulating inhibitor of coagulation, acquired thrombocyte dysfunction, and endotheliopathy in the pathogenesis of severe Lassa fever related coagulopathy.
Furthermore, in Crim Congo hemorrhagic fever, it has been shown that viscoelastic coagulation tests can differentiate coagulopathy that is mostly platelet dependent from hyperfibrinolysis or lack of coagulation factors.
Preliminary work from our group suggests that coagulation disorder in terms of slowed clot formation is noticeable even in mild cases by rotational thromboelastometry. Furthermore, patients with an adequate, strong inflammatory response without signs of liver failure or severe organ damage seem to preserve clot formation capacity in terms of maximum amplitude. Longitudinal measurements also reveal recovery of clot formation performance over time in LF survivors.
For the clinical management of this hemorrhagic fever with pandemic potential it would be crucial to know how exactly Lassa fever impairs coagulation and by which pathways it does so. Detailed understanding on this would allow clinicians to decide blood product management and to design host-directed therapies in severe Lassa fever in the future.
Hypothesis:
LF-linked coagulation disorder is present in mild and severe cases, revealed by advanced coagulation tests. LF coagulopathy is linked to endothelial dysfunction and is accompanied by clinical complications of vascular leak.
In aim 1, we will recruit hospitalized Lassa fever patients during 3 outbreak seasons that show clinical sings of hemorrhage or sonographic signs of vascular leakage, i.e. pleural or pericardial effusion, ascites, or evidence of pulmonary edema. Additionally, we will recruit randomly selected controls without hemorrhagic symptoms and sings of vascular leakage matched by sex and age.
Patients will receive bidaily clinical examinations, ultrasound exams, and basic laboratory, including inflammatory parameters, full blood count, liver and kidney function tests, RT-PCR for Lassa virus RNA, rotational thromboelastometry, and fibrinogen.
Additionally, a sub-cohort of hospitalized patients with Lassa fever complicated by acute kidney injury necessitating intermittent hemodialysis will receive coagulation assessment by thromboelastometry (ROTEM®) along with standard global coagulation tests (INR, aPTT) before and after hemodialysis. In this exploratory sub-cohort, the relationship between coagulation disorder detectable by standard vs. thromboelastometric tests and heparinization during dialysis and outcomes will be assessed.
Inactivated aliquots of the patients` samples will then be shipped to BNITM for assessment of various markers of coagulation, endothelial dysfunction, and platelet activation for aim 2 by multiplex bead-based immunoassays. Furthermore, a transcriptomic panel from peripheral blood mononuclear cells will allow us to integrate extensive biomarker data with clinical information on coagulation performance and formulate a working hypothesis on how exactly LASV causes coagulopathy in humans.
In aim 3, we plan to gather and streamline the laboratory and clinical information gained in this observational sub-study, to enhance current clinical care guidelines for the management of hospitalized LF. We will elaborate on what diagnostic tests are to be prioritized for LF patients at risk for hemorrhagic complications. Additionally, we will derive recommendations for guiding interventions in Lassa fever with risk of hemorrhagic complications and on coagulation management with blood products.
Project-related publications:
Blaise Lafoux, Nicolas Baillet, Caroline Picard, Gustave Fourcaud, Virginie Borges-Cardoso, et al. Hemostasis defects underlying the hemorrhagic syndrome caused by Mammarenaviruses in a cynomolgus macaque model. Blood, 2023, 142 (24), pp.2092-2104. doi: 10.1182/blood.2023020351.pasteur-04266945
Fletcher TE, Leblebicioglu H, Bozkurt I, Sunbul M, Bilek H, Asik Z, Barut S, Gunes F, Gemici U, Hewson R, Wilson D, O'Shea MK, Woolley T, Faragher B, Parmar K, Lalloo DG, Beeching NJ, Hunt BJ. Rotational thromboelastometry alongside conventional coagulation testing in patients with Crimean-Congo haemorrhagic fever: an observational cohort study. Lancet Infect Dis. 2019 Aug;19(8):862-871. doi: 10.1016/S1473-3099(19)30112-4. Epub 2019 Jun 28. PMID: 31262565; PMCID: PMC7641897.
-
Leonie Dreher (20. CS)
Role of the mineralocorticoid receptor in inflammatory cells in bacterial infection
Project leader: Prof. Dr. Ulrich Wenzel and Prof. Dr. Hans Willi Mittrücker
Affiliation: III. Department of Medicine and Department of Immunology
Background and preliminary data:
Since the introduction of non-steroidal mineralocorticoid receptor (MR) antagonists and novel aldosterone synthase inhibitors, interest in the aldosterone system has significantly increased (Lother 2022). Recent studies have shown that the MR in myeloid and lymphocytic cells plays a pro-inflammatory and pro-fibrotic role as we recently summarized (van der Heijden 2022, Hengel 2022). Using a CD11cCre×MRfl/fl and a CD4Cre×MRfl/fl mouse models, we and others found that inactivation of the MR in CD11c+ dendritic or CD4+ T cells reduces blood pressure and end-organ damage in arterial hypertension (Hengel 2022).
Does the lack of the MR in dendritic cells worsen bacterial infection?
The MR acts as a pro-inflammatory factor in myeloid and lymphocytic cells (Hengel 2022). Deficiency of the MR in inflammatory cells has been shown to be beneficial in autoimmune and cardiovascular disease (van der Heijden 2022). However, there is a lack of data on the role of the MR in inflammatory cells during bacterial infections. If the MR promotes inflammation in dendritic and CD4+ T cells, its inhibition could potentially be detrimental during infections, thus making its therapeutic targeting in humans impractical.
To investigate this crucial question, we will combine the CD11cCre×MRfl/fl and CD4Cre×MRfl/fl mouse models with two established infection models involving the kidney, spleen and liver. Systemic infection with Listeria monocytogenes induces a TH1 and CD8+ T cell response, while infection with Staphylococcus aureus induces a TH17 cell response (Krebs 2020, Bertram 2023). We will examine both the innate and adaptive immune phases. Key parameters will include bacterial counts in infected organs, kidney function, and histological analysis. Additionally, we will study the CD4+ and CD8+ T cell response to the pathogens. Infection with ovalbumin-recombinant listeria will allow us to assess antigen-specific T cell responses (listeriolysin O189-201 and ovalbumin257-264).
We have observed increased salt concentration in the skin in hypertension and knockout of the MR in dendritic cells leads to a significant reduction in this concentration. A high-salt diet results in increased salt storage in the skin and enhanced infection defense against cutaneous infection (Hengel 2022). Therefore, we will investigate whether the MR knockout in dendritic cells leads to reduced defense in cutaneous staphylococcal infection.
For effective activation, T cells require recognition of peptide MHC complexes by their T cell receptor and co-signals provided by CD28 ligands on the antigen presenting cell. Following primary activation, T cells differentiate into specialized effector cells. These process is primarily controlled by soluble factors, such as cytokines, present in the local environment or secreted by the antigen presenting cells. We will examine whether MR signaling affects T cell activation and differentiation either directly or by altering the function of antigen presenting cells during bacterial infection.
Hypothesis:
The MR in dendritic and lymphocytic cells plays an important role in control of bacterial infection.
Aims and Work Program:
1. Lack of the MR in dendritic cells or CD4+ T cells aggravates bacterial infection.
2. Lack of the MR in CD4+ T cells reduces the antigen-specific T cell response in listeria infection.
3. Overexpression of MR in dendritic or CD4+ T cells shows increased resistance to infections.
Aim #1 Infection with Listeria monocytogenes and Staphylococcus aureus
Listeria: Intravenous infection with Listeria monocytogenes (strain LmOVA) will be performed in CD11cCre×MRfl/fl and CD4Cre×MRfl/fl mice. For studying the innate immune response, organs will be harvested 2 days post-infection, and for the adaptive immune response, on day 10. The bacterial dose will be 5×104 for early examination and 5×103 for late examination. On day 2, bacterial titers in the liver, spleen, and kidneys will be determined, and tissues will be collected for histology. Neutrophil and inflammatory monocyte mobilization will be analyzed using FACS. Bacterial titers in the liver, spleen, and kidneys will be quantified by plating serial dilutions of homogenized tissue, along with abscess counts in kidney tissue. The accumulation of myeloid cells and T cells in the spleen, kidneys, and liver will be determined using FACS and immunohistology (Krebs 2020).
Staphylococcus: Infection and analyses will be largely similar to those for Listeria infection. The intravenous dose of Staphylococcus aureus (strain SH1000) will be 1×107 bacteria (Bertram 2023). Parameters such as blood pressure, albuminuria, and GFR will be assessed. In the kidneys, histology and gene expression of markers of renal injury will be evaluated. For skin infection, an intradermal dose of 1×106 bacteria will be administered to the flank. In addition to the analyses already described, daily measurement of lesion size and histological examination of the infected skin area at the end of the experiment will be performed. Since no immunodominant peptides are available for S. aureus, T cells will be polyclonally stimulated to determine the general induction of TH1 and TH17 cells. Cells from the spleen, kidneys, and draining lymph nodes (in the case of skin infection) will be isolated and incubated with PMA and ionomycin in the presence of Brefeldin A, and cytokines will be measured by FACS.
Aim #2: Antigen-specific T cell response
For LmOVA, immunodominant epitopes in listeriolysin O for CD4+ T cells and in ovalbumin for CD8+ T cells are known, allowing the analysis of the strength and cytokine profile of specific T cell responses and thus the role of the MR in T cell activation and differentiation. Spleen cells will be incubated in vitro with ovalbumin257−264 and listeriolysin O189−201 peptides for 4 in the presence of Brefeldin A, and cytokines IFN-γ, TNF-α, and IL-17A, as well as CD40L expression in T cells, will be measured by FACS.
Aim #3: Overexpression of MR in myeloid and lymphocytic cells
Finally, we will examine whether mice with targeted overexpression of MR in dendritic cells or CD4+ T cells exhibit increased resistance to infections caused by Listeria and Staphylococcus aureus.
In conclusion, we believe that this project will provide valuable new insights into the role of the MR in myeloid and lymphocytic cells during bacterial infections. By understanding the mechanisms underlying this process, we could pave the way for innovative treatments in cardiovascular and infectious diseases.
Project-related publications:
Bertram T, Reimers D, Lory NC, Schmidt C, Schmid J, C Heinig L, Bradtke P, Rattay G, Zielinski S, Hellmig M, Bartsch P, Rohde H, Nuñez S, Rosemblatt MV, Bono MR, Gagliani N, Sandrock I, Panzer U, Krebs CF, Meyer-Schwesinger C, Prinz I, Mittrücker H-W. Kidney-resident innate-like memory γδ T cells control chronic Staphylococcus aureus infection of mice. Proc Natl Acad Sci U S A. 2023, 120:e2210490120.
Hengel F, Benitah JP, Wenzel UO. Mosaic theory revisited: inflammation and salt play a central role in arterial hypertension. Cell Mol Immunol. 2022, 19:561-576.
Krebs CF, Reimers D, Zhao Y, Paust HJ, Bartsch P, Nuñez S, Rosemblatt MV, Hellmig M, Kilian C, Borchers A, Enk LUB, Zinke M, Becker M, Schmid J, Klinge S, Wong MN, Puelles VG, Schmidt C, Bertram T, Stumpf N, Hoxha E, Meyer-Schwesinger C, Lindenmeyer MT, Cohen CD, Rink M, Kurts C, Franzenburg S, Koch-Nolte F, Turner JE, Riedel JH, Huber S, Gagliani N, Huber TB, Wiech T, Rohde H, Bono MR, Bonn S, Panzer U, Mittrücker H-W. Pathogen-induced tissue-resident memory TH17 (TRM17) cells amplify autoimmune kidney disease. Sci. Immunol. 2020, 5:eaba4163.
Lother A, Jaisser F, Wenzel UO. Emerging fields for therapeutic targeting of the aldosterone-MR signaling pathway. Br J Pharmacol. 2022, 179: 3099-3102. van der Heijden CDCC, Bode M, Riksen NP, Wenzel UO. The role of the MR in immune cells in cardiovascular disease. Br J Pharmacol. 2022, 179:3135-3151.
Funding: Wenzel: WE 1688/19-3; Mittrücker: SFB1192 project A4, SFB1328 project A3.
-
Jack Winneberger (21. CS)
Endothelial targets of Blood-Brain Barrier Breakdown in bacterial Meningitis
Project leader: Prof. Dr. med. Tim Magnus, Dr. Peter Ludewig
Affiliation: Department of Neurology, University Medical Center Hamburg-Eppendorf
Background and preliminary data:
Bacterial meningitis is a severe infectious disease most commonly caused by Streptococcuspneumoniae, Neisseria meningitidis and Hemophilus influenzae. Despite receding incidencedue to advanced vaccine-development and roll-out, infections still lead to high rates of mortalityand lasting disability worldwide (1). Blood-Brain Barrier (BBB) Breakdown is a majorpathological hallmark involved in meningitis and leads to increased influx of bacteria, immunecells and vasogenic edema and subsequently poses the risk of brain herniation with fatalconsequences (1). As such, the discovery of targets that limit BBB breakdown could offer greatbenefit as adjunctive therapy besides antimicrobial agents. In our laboratory, we have identifiedseveral key mechanisms responsible for the breakdown of the blood-brain barrier (2, 3).Building on these findings, we now aim to elucidate how these processes drive blood-brainbarrier disruption in the context of meningitis. Understanding the precise molecular and cellularevents involved will inform novel therapeutic approaches to mitigate or prevent the devastatingneurological outcomes associated with this condition.
G-Protein coupled receptors (GPCR) are the largest mammalian receptor family. They areabundantly expressed on brain endothelial cells and have been shown to influence BBBintegrity (4). Furthermore, their therapeutic potential is underlined by the fact that roughly 30%of marketed drugs target a GPCR. GPCRs couple to specific and sometimes multiple subunitsof G-Proteins (Gαs, Gαi, Gαq/11, and Gα12/13) that mediate their intracellular effects. To studytheir effects on the BBB we have developed a mouse model that allows the activation ofGαq/11 specifically in brain endothelial cells using Designer Receptors Exclusively Activatedby Designer Drugs (DREADD). DREADD are mutated GPCRs that are specific for a G-Proteinsubunit and can only be activated by manufactured ligands (5). Expressing the Gαq/11-DREADD under a brain endothelial specific promotor (Slco1c1-CreERT2) allows us to study itsrole in a cell and time-specific manner. Preliminary results have shown that activation ofendothelial Gαq/11 leads to a rapid BBB opening and lethality after 90 minutes, similar to theclinical presentation seen in patients with pneumococcal meningitis. We aim to build upon thisnovel finding and further investigate the mechanisms of endothelial Gαq-dependent BBBbreakdown using our transgenic mouse line, establish potential therapeutic targets and testthese in a murine model of bacterial meningitis.
Hypothesis:
We hypothesize that Gq-dependent signaling pathways in endothelial cells play an importantrole in BBB breakdown following bacterial meningitis.
Aims and Work Programme:
1. We will use the Slco1c1-CreERT2x Gαq/11-DREADD mouse to characterize endothelialGαq/11-dependent BBB breakdown
2. Identifying Gαq/11-dependent targets in a murine model of bacterial meningitis
3. Integrating the results from Aims 1 and 2 to identify novel endothelial targets formodulation of BBB breakdown and test these in our meningitis model
Aim #1: Characterizing Slco1c1-CreERT2x Gαq/11-DREADD mouse
We have established the Slco1c1-CreERT2x Gαq/11-DREADD mouse line in our lab allowingus to specifically activate Gaq-signaling in a brain endothelial- and time-specific manner.Preliminary results have shown that activation of these DREADD lead to a rapid neurologicaldeterioration and BBB breakdown as assessed by Evans Blue dye extravasation, indicating aprominent and so far poorly understood mechanism of BBB damage. To characterize thisobservation, we will use MRI including Perfusion imaging to assess differences in cerebralblood flow and edema formation. Furthermore, histological assessment and Western Blottingof endothelial junction proteins will be performed to assess the parenchymal injury. We havealso established a brain endothelial cell line (bEND.3) stably transfected with the Gαq/11-DREADD construct to further assess barrier properties via electrical cell-substrate impedancesensing (ECIS) and cytochemistry. Performing RNA Sequencing of these activated endothelialcells at different time-points will help us identify downstream targets that mediate this BBBbreakdown. To validate these results in vivo, we will use an established protocol to isolateendothelial nuclei for single-nucleus RNA-sequencing.
Aim #2: Identifying Gαq/11-dependent targets in a murine model of bacterialmeningitis
We will establish a murine model of streptococcal meningitis as previously described (1). Inbrief, meningitis is induced by transcutaneous injection of 15 μl 107 colony forming units(cfu)/ml of Streptococcus pneumoniae (S. pneumoniae) type 3 into the cisterna magna.Performing single-nucleus RNA-sequencing of endothelial cells and correlating this with thesequencing results of our endothelial cells from Aim #1 will allow us to uncover preservedGαq/11-dependent targets that are upregulated in experimental meningitis.
Aim #3: Isolating new therapeutical targets to limit BBB breakdown in bacterialmeningitis
Building upon the potential endothelial candidate genes identified in Aim #1 and #2 we will firstvalidate therapeutic targets in vitro using our Gq-DREADD endothelial cell line and ECISmeasurements as a model of endothelial barrier breakdown. In a second step, promisingtargets will be tested in our murine model of bacterial meningitis. Outcomes will include clinicalscore, edema formation, CSF pleocytosis and ICP-measurements.In conclusion, we believe that Gαq/11-dependent signaling in brain endothelial cells is a vitalmediator of BBB breakdown. A better understanding of this process and its role in bacterialmeningitis will help identify valuable therapeutic targets to limit the consequences of excessivebrain edema as adjunctive therapy to antimicrobial treatment.
Project-related publications:
1. Teske, N. C., Dyckhoff-Shen, S., Beckenbauer, P., Bewersdorf, J. P., Engelen-Lee, J.-Y.,Hammerschmidt, S., Kälin, R. E., Pfister, H.-W., Brouwer, M. C., Klein, M., Glass, R., van deBeek, D., and Koedel, U. (2023) Pericytes are protective in experimental pneumococcalmeningitis through regulating leukocyte infiltration and blood–brain barrier function. Journalof Neuroinflammation 20, 267
2. Ludewig, P., Sedlacik, J., Gelderblom, M., Bernreuther, C., Korkusuz, Y., Wagener, C., Gerloff,C., Fiehler, J., Magnus, T., and Horst, A. K. (2013) Carcinoembryonic antigen-related celladhesion molecule 1 inhibits MMP-9-mediated blood-brain-barrier breakdown in a mousemodel for ischemic stroke. Circ Res 113, 1013-1022
3. Winneberger, J., Schols, S., Lessmann, K., Randez-Garbayo, J., Bauer, A. T., Mohamud Yusuf,A., Hermann, D. M., Gunzer, M., Schneider, S. W., Fiehler, J., Gerloff, C., Gelderblom, M.,Ludewig, P., and Magnus, T. (2021) Platelet endothelial cell adhesion molecule-1 is agatekeeper of neutrophil transendothelial migration in ischemic stroke. Brain Behav Immun93, 277-287
4. B S, G., B S, N., K, R., C S, M., and R, S. (2024) Unlocking the therapeutic capabilities of GPCRin the treatment of ischemic stroke: A translational literature. Medicine in Drug Discovery 24,100197
5. Claes, M., De Groef, L., and Moons, L. (2022) The DREADDful Hurdles and Opportunities ofthe Chronic Chemogenetic Toolbox. Cells 11
-
Annerose Ziegler (22. CS)
TBD
-
Sonia-Emilia Selicean (23. CS)
Stage- and disease-specific signatures of immune dysfunction indecompensating advanced chronic liver disease
Project leader: Priv.-Doz. Dr. med. Peter Huebener
Affiliation: I. Medical Department / University Liver Center, UKE
Background and preliminary data:
The liver plays crucial roles in metabolism, detoxification andimmunity. Advanced chronic liver disease (ACLD), the final stage of various hepatic diseases,encompasses functional and microarchitectural alterations to precipitate tissue dysfunction andclinical complications. As such, acute-on-chronic liver failure (ACLF), a frequent and devastatingcomplication of ACLD, comprises hepatic and/or extrahepatic organ failure, associates withsignificant morbidity and mortality, and currently lacks targeted treatment options (1). The syndromeis most often triggered and/or complicated by bacterial infections, highlighting the central role of theliver in systemic immunity (2). Importantly, ACLF is reversible in a significant fraction of patients,indicating “molecular reprogramming” potentially amenable for targeted interventions. To studymechanisms of ACLF-associated immune dysfunction may thus not only deepen our understandingof the liver in host defense, but may also reveal targetable molecular pathways to stabilize or evenrevert ACLD manifestations, including susceptibility to infections.
Multi-omics approaches including transcriptomics, proteomics, and advanced imaging can be usedfor the generation of multidimensional signatures of disease states (3). Integration of thesetechniques constitutes a promising approach for a comprehensive characterization of molecularchanges during ACLD, as well as for the development of strategies to prevent, delay or even revert the need for LT.
Hypothesis:
Clinical ACLD stages are characterized by disease- and stage-specific alterations inliver microarchitecture as well as transcriptional and translational signatures, which affect immunenetworks to facilitate infections and dysregulate host responses to pathogens. Investigations intothe molecular foundations of ACLD stages may aid in the identification of signaling pathwaysassociated with disease progression and loss of liver immunologic functionality. The anticipatedfindings may inform strategies to reprogram the failing liver into an at least temporarily stable state,and to revert cirrhosis-associated immune dysfunction to prevent infections and infection-relatedcomplications.
Aims and Work Programme:
Aim 1: To characterize stage-specific tissue architectural, transcriptional and translationalalterations in human liver tissues across a spectrum of etiologically diverse chronic liver diseases(i.e., alcoholic liver disease “ALD”, chronic hepatitis C infection “HCV”, primary sclerosing cholangitis“PSC”) at different stages of decompensation (ACLD to ACLF) with a predominant focus on immunecell architecture and immune signaling networks.
Work programme: We will use 8-10 samples per disease stage (healthy resection margins from livermetastasectomies, “HRM” as controls, CC, DC, ACLF) each from three etiologically diverse chronicliver diseases (ALD, HCV, PSC) to decipher commonalities and disease-specific ACLD-stagesignatures. Samples will processed via bulk transcriptomics and whole tissue proteomics.In preparation of this proposal, we have identified above mentioned samples in our tissue biobanks(patient consent available), which may be used upon funding approval. Selected representativesamples will additionally be used for single cell sequencing, but these will need to be prospectivelyharvested during LT in a protocol-based manner. A cooperation with the Department of Visceral Transplant Surgery (UKE) has been informally established in preparation of this proposal.
Aim 2: To characterize immune microarchitectural changes associated with ACLD progression.
Work programme: samples from the above mentioned explants will be processed for spatialtranscriptomics and multiplex immunofluorescence imaging, using established cell and liver zonationmarkers, as well as known or newly described (Aim 1) functional markers relevant in regulating key(patho-)physiological pathways in liver parenchyma. Multiplex-IF will aid in the mapping of signalingalterations to liver parenchymal and stromal cells as well as to liver-resident immune cells includingKupfer cells, hepatocytes, stellate cells and others. Bioinformatic analysis will aid in qualitative andquantitative assessment of disturbances in lobular and sublobular liver architecture and functionality.
Aim 3: To assess the functional and phenotypical validity of current and newly developed rodentmodels of ACLD as scientific tools for the identification of relevant biological pathways and pretestingof medical treatments for different stages of ACLD and particularly infectious complications.
Work program: Liver tissues from rodent models, including the “chronic CCl4 model”, the Mdr2-/-model of progressive cholestatic liver disease as well as the Tak1Δhep model of chronic cell deathdependenthepatopathy will be compared to human liver tissues samples from aims 1 and 2 withregards to their phenotypic and (dys-)functional similarities to human ACLD and particularly ACLF.In subsequent studies, LPS and infections with live bacteria will elicit functional consequences ofACLD-associated dysregulated host responses to pathogens.
References
1. Arroyo V, Moreau R, Jalan R. Acute-on-Chronic Liver Failure. N Engl J Med. 2020 May 28;382(22):2137–45.
2. Albillos A, Martin-Mateos R, Van der Merwe S, Wiest R, Jalan R, Álvarez-Mon M. Cirrhosis-associatedimmune dysfunction. Nat Rev Gastroenterol Hepatol. 2022 Feb;19(2):112–34.
3. Kreitmaier P, Katsoula G, Zeggini E. Insights from multi-omics integration in complex disease primarytissues. Trends Genet TIG. 2023 Jan;39(1):46–58.
-
Chiara Cattaneo (24. CS)
TBD
-
Aiman Gamal Abdelrahim (25. CS)
Innate and adaptive immune control of Mpox virus infection
Project leader: Prof. Dr. Marylyn Addo and Dr. Dr. Angelique Hölzemer
Affiliation: I. Department of Medicine and IIRVD
Background and preliminary data:
Mpox virus (MPXV) is a double stranded DNA-virus of the genus orthopoxvirus that was historically confined to Africa but has continued to spread globally since 2022. This led to the declaration of two public health emergencies of international concern, the latest starting on 14 August 2024 by the World Health Organization (WHO) (1). As of November 2024, there have been over 117,000 cases with 263 confirmed deaths worldwide (2). In endemic areas, pregnant women, children and immunosuppressed individuals (including PLHIV), are at risk for severe clinical disease. In non-endemic areas groups at high risk for MPXV infection are men who have sex with men and people living with HIV-1 (PLHIV). How HIV-1 infection (treated or untreated) affects an individual’s risk for severe disease or viral spread of MPXV is at present not fully understood.
MPXV primarily infects human skin cells, particularly keratinocytes, but can also invade other tissues such as lymph nodes, liver, and spleen. Innate immune cells play key roles in the early immune response to MPXV infection and mice with low natural killer (NK) cell numbers are highly vulnerable to lethal MPXV infection (3,4) There are two primary clades of MPXV: the West African clade II, responsible for the current global outbreak, and the more virulent Central African clade I. The difference in the human NK cell response to these two different clades and how that influences virulence is currently unknown. This project aims to uncover how NK cells as innate effector cells react to and control MPXV infection, and whether the interaction between NK cells and infected target cells is dysfunctional in PLHIV.
Hypothesis:
The central hypothesis of this project is that NK cells kill MPXV-infected target cells via activating receptor-ligand interactions. Both immune-evasive MPVX proteins and NK-cell dysfunction can result in ineffective recognition of MPXV-infected targets by NK cells.
Aims and Work Program:
1. To characterise NK-cell receptor ligands differentially surface expressed on MPXV-infected epithelial cells, monocytes and hepatocytes.
2. To determine the NK-cell response against MPXV-infected targets in people living with HIV-1, healthy controls and individuals vaccinated with modified vaccinia virus Ankara (MVA vaccine against smallpox).
In Aim #1, we will analyse how MPXV infection modulates the surface expression of NK-cell receptor ligands on infected target cells using an established flow cytometry panel (5) and intracellular staining with an anti-nucleoprotein antibody against poxviruses. For MPXV infection, we will use epithelial cell lines (i.e. A549 cells), primary hepatocytes and autologous primary cells (i.e. monocytes/macrophages) as target cells and compare Clade Ia, Ib and Clade II MPXV strains. Our collaborator recently showed that Sars-CoV2 infection induced shedding of MIC-A and B molecules from the cell surface of infected A549 cells (6), which was reversed by using the monoclonal antibody 7C6. This reduced NK-cell killing of infected cells by preventing engagement of the activating NKG2D receptor, which has also been implicated in NK-cell control of poxvirus disease (7,8). Here, we aim to investigate, whether MPXV employs similar (potentially targetable) mechanisms to subvert NK-cell immunity. These studies will be done under S3 biosafety conditions, additionally training the ID Fellow candidate to perform experiments with viruses of biosafety level 3.
In Aim #2, we will investigate whether NK cells are able to prevent spread of MPXV infection in cell culture. Next to direct NK-cell killing of MPXV infected target cells, we will measure the production of antiviral cytokines such as Type II interferons in response to co-culture with MPXV-infected targets. We will compare the antiviral and cytotoxic activity of NK cells derived from peripheral blood from donors without HIV-1, with HIV-1 and of donors which have received one or two doses of the MVA vaccine against smallpox. This vaccine provides adaptive heterologous immunity to MPXV, and this work will investigate whether there is an additional effect of trained innate immunity next to adaptive immune control in vaccinated individuals. Additionally, the extent of persistent NK-cell dysfunction in treated PLHIV against MPXV will be investigated in vitro. Ethics approval has been obtained. Patient and vaccine samples have already been obtained, and recruitment is ongoing.
Project-related publications:
1. https://www.who.int/news/item/14-08-2024-who-director-general-declares-mpox-outbreak-a-public-health-emergency-of-international-concern (2024).
2. World Health Organization. Mpox: Multi-country External Situation Report no. 44, published 23 December 2024. WHO (2024).
3. Earl, P. L., Americo, J. L. & Moss, B. Insufficient Innate Immunity Contributes to the Susceptibility of the Castaneous Mouse to Orthopoxvirus Infection. J. Virol. 91, 1–17 (2017).
4. Earl, P. L., Americo, J. L. & Moss, B. Natural killer cells expanded in vivo or ex vivo with IL-15 overcomes the inherent susceptibility of CAST mice to lethal infection with orthopoxviruses. PLoS Pathog. 16, 1–25 (2020).
* 5. van Stigt Thans, T., Akko, J. I., Niehrs, A., Garcia-Beltran, W. F., Richert, L., Stürzel, C. M., Ford, C. T., Li, H., Ochsenbauer, C., Kappes, J. C., Hahn, B. H., Kirchhoff, F., Martrus, G., Sauter, D., Altfeld, M. & Hölzemer, A. Primary HIV-1 Strains Use Nef To Downmodulate HLA-E Surface Expression. J. Virol. 93, 10.1128/jvi.00719-19 (2019).
* 6. Hartmann, J. A., Cardoso, M. R., Talarico, M. C. R., Kenney, D. J., Leone, M. R., Reese, D. C., Turcinovic, J., O'Connell, A. K., Gertje, H. P., Marino, C., Ojeda, P. E., De Paula, E. V., Orsi, F. A., Velloso, L. A., Cafiero, T. R., Connor, J. H., Ploss, A., Hoelzemer, A., Carrington, M., Barczak, A. K., Crossland, N. A., Douam, F., Boucau, J. & Garcia-Beltran, W. F. Evasion of NKG2D-mediated cytotoxic immunity by sarbecoviruses. Cell 187, 2393–2410.e14 (2024).
7. Fang, M., Lanier, L. L. & Sigal, L. J. A role for NKG2D in NK cell-mediated resistance to poxvirus disease. PLoS Pathog. 4, (2008). 8. Campbell, J. A., Trossman, D. S., Yokoyama, W. M. & Carayannopoulos, L. N. Zoonotic orthopoxviruses encode a high-affinity antagonist of NKG2D. J. Exp. Med. 204, 1311–1317 (2007).
-
Katharina Moll (26. CS)
Elucidating the role of Interleukin 34 during bacterial infections
Project leader: Prof. Dr. Samuel Huber and Prof. Dr. Hans-Willi Mittrücker
Affiliation: I. Medizinische Klinik und Poliklinik and Institut für ImmunologieUniversitätsklinikum Hamburg-Eppendorf
Background and preliminary data:
Inflammation is fundamental in the host’s defence against invading pathogens. Not only the type ofpathogen and its equipment of virulence factors determine severity of the infection but also host factorscan aggravate or ameliorate the course of disease. Recently, interleukin 34 (IL-34) has been identifiedas the second ligand for the Colony Stimulating Factor 1 receptor (CSF-1R). Interestingly, IL-34 seemsto play a crucial role in the differentiation and proliferation of macrophages, which express CSF-1R.Previous studies further indicate that IL-34 is upregulated in various infectious diseases includingbacterial infections. However, these studies were mostly based on correlative analyses and in vitroexperiments. Functional in vivo experiments have not been performed so far.
Therefore, in a first step, we analyzed the IL-34 expression in mice under steady state conditions andin models of local inflammation. We found that IL-34 is expressed in various organs and by many celltypes such as dendritic cells, neutrophils, granulocytes and macrophages during homeostasis.However, during inflammation, non-hematopoietic cells seem to be the most relevant source. In asecond step, we analyzed IL-34 expression in mice upon infection with L. monocytogenes and foundupregulation of IL-34 in spleen and liver. However, functional experiments addressing the relevance ofthis finding have not been done so far. Thus, and to study the role of IL-34 on a functional level, we havegenerated and validated Il34-deficient mice.
Hypothesis:
Based on our preliminary data and the published literature, we hypothesize that non-immune cell derivedIL-34 plays a protective role in bacterial infections by acting on macrophages.
Aims and Work Programme:
We want to elucidate the role of IL-34 in the pathogenesis of bacterial infections. To this end, we willuse Il34-deficient mice and mouse models for bacterial infections (e.g. L. monocytogenes,Staphylococcus aureus, Salmonella typhimurium, Citrobacter rodentium). First, we will measure theexpression level and cellular source of IL-34 in infection. Second, we will use Il34-deficient mice to studythe functional role of IL-34 in these infections. We will assess disease severity and control of the bacteria.We will further evaluate innate and adaptive immune response of infected mice in terms of cellularphenotype and cytokine profile of immune cells using flow cytometry. We will use established sets ofantibodies to determine the response of different cell types (epithelial cells, T and B cells as wellmacrophages and dendritic cells). Additionally, we will perform an unsupervised approach by CITEseqof total cells isolated from colon of wildtype mice with or without infection. With this strategy, we minimizea potential bias due to purification protocols designed for specific cell population. Third, we aim to identifythe target cells of IL-34. Based on our preliminary data, B cells and macrophages are potential IL-34targets. To this end, we will test the in vitro response of defined cell subsets to IL-34 stimulation. We willanalyse the proliferation and cytokine profile of these cells. Furthermore, as CSF-1R is the knownreceptor of IL-34, we have crossed Csf1r conditional knockout mice with LysM-cre (macrophage)and CD19-cre (B cell) mice in order to identify relevant target cells of IL-34. Finally, we aim to translateour findings to human biology. Thus, we will measure IL-34 levels in patients during the course ofbacterial infections and correlate its expression with clinical parameters. Taken together, this study willidentify the role of IL-34 in infection and thereby build the basis for future immunotherapies designed ontargeting this cytokine.
Project-related publications:
1. Pelczar P, Witkowski M, Perez LG, Kempski J, Hammel AG, Brockmann L, Kleinschmidt D,Wende S, Haueis C, Bedke T, Witkowski M, Krasemann S, Steurer S, Booth CJ, Busch P, KönigA, Rauch U, Benten D, Izbicki JR, Rösch T, Lohse AW, Strowig T, Gagliani N, Flavell RA, HuberS. A pathogenic role for T cell-derived IL-22BP in inflammatory bowel disease. Science.2016;354(6310):358-62.
2. Baghdadi M, Umeyama Y, Hama N, Kobayashi T, Han N, Wada H, Seino KI. Interleukin-34, acomprehensive review. J Leukoc Biol. 2018 Nov;104(5):931-951.
3. Bartsch P, Kilian C, Hellmig M, Paust HJ, Borchers A, Sivayoganathan A, Enk L, Zhao Y, ShaikhN, Büttner H, Wong MN, Puelles VG, Wiech T, Flavell R, Huber TB, Turner JE, Bonn S, HuberS, Gagliani N, Mittrücker HW, Rohde H, Panzer U, Krebs CF. Th17 cell plasticity towards a Tbet-dependent Th1 phenotype is required for bacterial control in Staphylococcus aureusinfection. PLoS Pathog. 2022 Apr 21;18(4):e1010430.
4. Lücke K, Yan I, Krohn S, Volmari A, Klinge S, Schmid J, Schumacher V, Steinmetz OM, Rose-John S, Mittrücker H-W. Control of Listeria monocytogenes infection requires classical IL-6signaling in myeloid cells. PLoS One. 2018 Aug 31;13(8):e0203395.
5. Hoenow S, Yan K, Noll J, Groneberg M, Casar C, Lory NC, Vogelsang M, Hansen C, Wolf V,Fehling H, Sellau J, Mittrücker H-W, Lotter H. The Properties of Proinflammatory Ly6ChiMonocytes Are Differentially Shaped by Parasitic and Bacterial Liver Infections. Cells. 2022 Aug16;11(16):2539.
-
Jakob Malsy (1. Associated member)
Innate immune response to human norovirus infection in human intestinal organoids
Project leader: Prof. Dr. Marcus Altfeld
Affiliation: Leibniz Institute of Virology
Background and preliminary data: Human Noroviruses (hNVs) are the leading cause of non-bacterial gastroenteritis worldwide and a major factor of diarrhea-associated deaths in children in developing countries. During each winter, up to 5000 cases of hNV infections per week are reported in Germany. Pathogenesis and immunologic responses to hNV infection remain largely unknown and no causal therapy is currently available. The lack of a robust experimental model for studying hNVs pathogenesis has hindered the development of effective therapeutic interventions. Recently, the landscape of hNV research has changed importantly with the initial demonstration in 2016 that hNVs can replicate in human intestinal organoids1 and the subsequent demonstration in July 2022 that hNV can also replicate in a primary salivary cell line2. These novel in vitro models will now enable to investigate hNV biology and innate host responses using human cell systems.
Pattern recognition receptors (PRRs) play a key role in the early immune response to viral infection, inducing innate Type I Interferon (IFN) responses and initiating subsequent adaptive immune responses. During murine NV infections, nucleotide-binding and oligomerization domain like receptor (NLR) activation and stimulation of the cGAS-STING-pathway have been described; however, the specific PRRs that can recognize hNV infection remain unknown. In Aims #1 and #2 of this project, human intestinal organoids and salivary cells will be used to identify the PRRs that sense hNV infection, and the mechanisms by which PRR sensing of hNV induces antiviral Type I IFN responses. Aim #3 will build on previous studies of the Altfeld lab demonstrating that antiviral Type I IFN responses differ between the sexes, with consequences for the clinical manifestations of viral infections3,4. Sex differences in hNV infection have also emerged from the case numbers reported to the Robert Koch Institute in Germany. While incidences of hNV infections are higher in male children, hNV incidences after puberty are higher in females. In Aim #3, we plan to investigate whether PRR sensing of hNV induces sex-specific differences in Type I IFN responses, and whether these differences are underlying the sex-differences in reported hNV cases and manifestations.
The Altfeld laboratory has extensive experience in investigating innate and adaptive immune responses against viral infections5, including the use of human intestinal organoid models. Preliminary studies have furthermore demonstrated successful hNV infection of human intestinal monolayers and indicated NLR activation (see below).
Left image: HNV Capsid (VP1) positive cells from human intestinal monolayers, identified using flow cytometry, after 1 hour past NV infection (blue) and 48 hours past infection (orange), indicating hNV replication. Right image: ASC speck formation in a human intestinal monolayers as a surrogate for NLR activation after incubation with hNV. Scale bar 20 μm.
Aims and Work-Programme:
Aim #1 – Identification of PRR expression pattern during the early immune response to hNV infection
Hypothesis: Specific PRR pathways are activated in human intestinal organoids during early hNV infection, through sensing of hNVs and/or cell damage.
Approach: Upon infection of human intestinal organoids (HIO) and infection of salivary primary cell lines, expression of PRRs and of key regulators of PRR signaling will be assessed, using qRT-PCR and multiparameter flow cytometry. Specifically, expression of TLR3-4, TLR7-9, NLRs, RIG-1, MDA5, cGAS, STING, P2X7, MyD88, Caspase-1, NFkB, IRF7 and IRF3 will be assessed during early and late acute infection.
Aim #2 – Assessing the consequences of PRR signaling during hNV infection
Hypothesis: PRR signaling influences infection rates and replication of hNVs in HIOs.
Approach: Induction of Type I IFNs and other antiviral cytokines (TNF, Il-1, Il-6, Il-18) will be quantified following infection. We will generate HIOs knockouts, using the CRISPR-Cas9 platform in our laboratory, to inactivate the specific PRRs identified in Aim 1 in order to determine their contribution to antiviral effector functions. Knockouts and naïve HIOs will be infected with hNVs and cell viability, organelle damage, infection rates and hNV replication will be assessed to identify effects of the specific PRRs during hNV infection.
Aim #3 – The role of sex hormones in regulating hNV infection and PRR signaling
Hypothesis: Sex hormones influence hNV infection through regulation of PRR expression.
Approach: To determine whether increased levels of estrogens, testosterone and/or progesterone influence hNV infection and viral replication of hNVs, HIOs will be incubated with the respective hormones before infection. We will investigate whether sex-hormones alter expression of PRRs identified in Aims 1 & 2, or whether they impact downstream signaling.
Project-related publications:
[1] Ettayebi, K., Crawford, S. E., Murakami, K., Broughman, J. R., Karandikar, U., Tenge, V. R., Neill, F. H., Blutt, S. E., Zeng, X.-L., Qu, L., Kou, B., Opekun, A. R., Burrin, D., Graham, D. Y., Ramani, S., Atmar, R. L., & Estes, M. K. (2016). Replication of human noroviruses in stem cell-derived human enteroids. Science, 353(6306), 1387–1393.
[2] Ghosh, S., Kumar, M., Santiana, M., Mishra, A., Zhang, M., Labayo, H., Chibly, A. M., Nakamura, H., Tanaka, T., Henderson, W., Lewis, E., Voss, O., Su, Y., Belkaid, Y., Chiorini, J. A., Hoffman, M. P., & Altan-Bonnet, N. (2022). Enteric viruses replicate in salivary glands and infect through saliva. Nature, 607.
[3] Hagen, S. H., Henseling, F., Hennesen, J., Savel, H., Delahaye, S., Richert, L., Ziegler, S. M., & Altfeld, M. (2020). Heterogeneous Escape from X Chromosome Inactivation Results in Sex Differences in Type I IFN Responses at the Single Human pDC Level. Cell Reports, 33(10).
[4] Ziegler, S. M., & Altfeld, M. (2017). Human immunodeficiency virus 1 and type I interferons-where sex makes a difference. Frontiers in Immunology, 8(Sep).
[5] Jung, J. M., Ching, W., Baumdick, M. E., Hofmann-Sieber, H., Bosse, J. B., Koyro, T., Möller, K. J., Wegner, L., Niehrs, A., Russu, K., Ohms, M., Zhang, W., Ehrhardt, A., Duisters, K., Spierings, E., Hölzemer, A., Körner, C., Jansen, S. A., Peine, S., … Bunders, M. J. (2021). KIR3DS1 directs NK cell–mediated protection against human adenovirus infections. Science Immunology, 6(63).
-
Jan Philipp Weltzsch (2. Associated member)
Biliary microbiota composition determines epithelial barrier function and risk of acutebacterial cholangitis
Project leader: Prof. Dr. med. Christoph Schramm and Victor Haas
Affiliation: I. Department of Medicine and Martin Zeitz Center for Rare Diseases
Background and preliminary data:
Acute bacterial cholangitis is an infection of the biliary tract, often as a consequence of biliarytract obstruction. Despite improved management, severe complications such as biliary sepsiscan arise, and the mortality associated with acute cholangitis remains around 5%1. Causalbacterial species mainly identified are Escherichia coli, Klebsiella spp and Enterococcus spp.People affected by Primary Sclerosing Cholangitis (PSC), a disease of unknown etiologyleading to scarring and fibrosis of the intrahepatic and extrahepatic bile ducts, are at high riskof developing acute bacterial cholangitis. We have shown using 16s rRNA sequencing frombile fluid, that even in the absence of acute bacterial cholangitis, the biliary tract is colonizedby a diverse bacterial ecosystem both in non-PSC controls and people suffering from PSC2.People with PSC display a lower microbial diversity in the bile ducts, accompanied by anexpansion of pathogens2 (Fig.1A). Notably, the opportunistic pathogen Enterococcus faecaliswas significantly increased in stool and bile samples of people with PSC, and the presence ofEnterococcus spp in bile associated with lower liver transplantation-free survival in PSC3,4(Fig.1B). We have already collected and sequenced the genome of 49 biliary E. faecalisisolates from people with PSC and non-PSC controls. Genomic analysis allowed us to identifyharmless and virulent E. faecalis strains, that harbor a vast array of virulence genes and displayhigh proinflammatory potential in vitro (Fig.1C).
Emerging data highlight the predictive value of microbiota composition in identifying patientsat risk of further infection. In liver transplant recipients, low gut microbial diversity associatedwith increased risk of postoperative infection, and high Enterococcus spp abundance had apredictive value in this setting5. Biliary microbes are in direct contact with cholangiocytes, thecells forming the lining of the bile ducts. Maintenance of the biliary epithelial barrier (BEB)integrity is crucial for pathogen defense and homeostasis in the liver, and we believe thatmicrobiota composition is critical to its functionality. By understanding host-microbe dynamicsat the BEB, we hope to further uncover the pathogenesis of acute bacterial cholangitis, and ona larger scale to assess the importance of the unexplored biliary microbiome in liver physiology.
Hypothesis:
We hypothesize that biliary dysbiosis and expansion of virulent Enterococcus sp in the bileleads to biliary barrier dysfunction and is thus a risk factor for acute bacterial cholangitis inPSC and post liver transplantation.
Aims and Work Programme:
1. To investigate the relationship between the biliary microbiome and occurrence of acutebacterial cholangitis in a well characterized cohort of people affected by PSC and postliver transplantation.
2. To investigate the potential of biliary commensals and pathogens to modulate thebiliary epithelial barrier function.
In Aim #1, we will rely on 16s rRNA sequencing analyses as well as our large and ongoingcollection and whole genome sequencing data of bile samples and biliary isolates. We will:
- Perform 16s rRNA sequencing of consecutive bile sample collected from our PSC and post-transplant cohort and analyze the stability of microbial communities over time andcourse of disease.
- Assess whether microbial patterns are predictive for the occurrence of acute bacterialcholangitis.
- Assess whether E. faecalis strain diversity in the bile i.e presence of harmless andvirulent strains is associated with higher risk of acute bacterial cholangitis.
- Investigate how antimicrobial therapies affect biliary microbial composition, and thuspotentially the risk of bacterial cholangitis
In Aim #2, we will investigate the crosstalk of biliary commensals and pathogens withcholangiocytes, ex vivo using PSC biosamples and in vitro using biliary organoid co-culturesystems. We plan to:
- Perform 16s rRNA sequencing of bile samples and mRNA sequencing of paired biliarybrushes collected during endoscopic procedures, to perform correlation analyses ofbiliary microbiota and cholangiocyte transcriptomic profiles.
- Challenge biliary organoids with live bacteria and assess the potential of these bacteriato induce changes in cholangiocyte barrier function, as assessed by transepithelialelectrical resistance measurements and production of antimicrobial peptides in theculture supernatant assessed by ELISA.
- Perform translocation assays to assess the capacity of biliary bacterial strains totranslocate across the biliary barrier.
Project-related publications:
(1) Sokal, A.; Sauvanet, A.; Fantin, B.; de Lastours, V. Acute Cholangitis: Diagnosis and Management. J Visc Surg 2019, 156 (6),515–525.
(2) Liwinski, T.; Zenouzi, R.; John, C.; Ehlken, H.; Rühlemann, M. C.; Bang, C.; Groth, S.; Lieb, W.; Kantowski, M.; Andersen, N.;Schachschal, G.; Karlsen, T. H.; Hov, J. R.; Rösch, T.; Lohse, A. W.; Heeren, J.; Franke, A.; Schramm, C. Alterations of theBile Microbiome in Primary Sclerosing Cholangitis. Gut 2020, 69 (4), 665–672.
(3) Zigmond, E.; Zecher, B. F.; Bartels, A.-L.; Ziv-Baran, T.; Rösch, T.; Schachschal, G.; Lohse, A. W.; Ehlken, H.; Schramm, C.Bile Duct Colonization With Enterococcus Sp. Associates With Disease Progression in Primary Sclerosing Cholangitis. ClinGastroenterol Hepatol 2023, 21 (5), 1223-1232.e3.
(4) Awoniyi M, Wang J, Ngo B, Meadows V, Tam J, Viswanathan A, Lai Y, Montgomery S, Farmer M, Kummen M, Thingholm L,Schramm C, Bang C, Franke A, Lu K, Zhou H, Bajaj JS, Hylemon PB, Ting J, Popov YV, Hov JR, Francis HL, Sartor RB.Protective and aggressive bacterial subsets and metabolites modify hepatobiliary inflammation and fibrosis in a murine modelof PSC. Gut 2023 Apr;72(4):671-685. doi: 10.1136/gutjnl-2021-326500.
(5) Lehmann, C. J.; Dylla, N. P.; Odenwald, M.; Nayak, R.; Khalid, M.; Boissiere, J.; Cantoral, J.; Adler, E.; Stutz, M. R.; Cruz, M.D.; Moran, A.; Lin, H.; Ramaswamy, R.; Sundararajan, A.; Sidebottom, A. M.; Little, J.; Pamer, E. G.; Aronsohn, A.; Fung, J.;Baker, T. B.; Kacha, A. Fecal Metabolite Profiling Identifies Liver Transplant Recipients at Risk for Postoperative Infection.Cell Host & Microbe 2024, 32 (1), 117-130.e4.