Signal Transduction Group (AG3)
PD Dr. Malte Kriegs, Dipl. Biochem.
Dr. Leoni Ramke, Clinician Scientist
Dr. Jacob F. Clausen, Clinician Scientist
Konstantin Hoffer, BTA
Eva Kinkel, BTA
Emily Schulz, Student
Contact: m.kriegs@uke.de
Aim:
The research group focuses on kinase-mediated signal transduction in cancer cells and the associated therapy-relevant processes such as cell proliferation, survival, radio- and chemo resistance.
We aim to identify hyperactive and therapy relevant kinases in head & neck cancer as well as glioblastoma to use them as targets for new therapeutic approaches. Possible resistance mechanisms are also being analyzed, such as the activation of kinases by X-rays. In addition to classical cell biology, biochemical and protein chemistry methods, the central method in our studies is the functional kinomprofiling. This is an array-based key technology for the functional analysis of the kinome. The kinome profiling is organized in our UCCH Kinomics Core Facility (link).
Specific topics:
UCCH Kinomics Core Facility
- Signal transduction in head & neck cancer (HNSCC)
Influence of kinase signaling on therapy sensitivity in Glioblastoma (GBM)
- Radiation induced kinase signaling
- Predictive marker for immune checkpoint inhibition
- Response prediction of kinase inhibitors in personalized cancer therapy
Core Project: UCCH Kinomics Core Facility
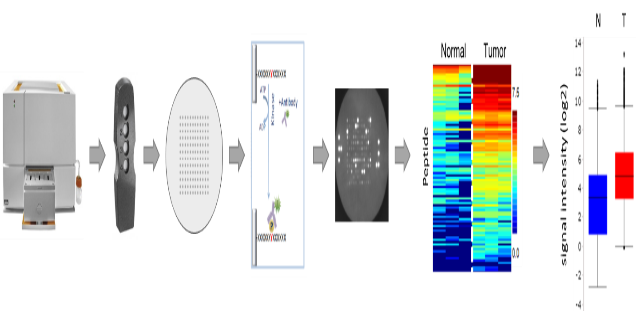
The UCCH Kinomics Core Facility uses a PamStation®12 from PamGene Int. B.V. to perform functional kinome profling, a key technology to analyze kinases on an omics level. This peptide array based system measures the activity of kinases in native protein lysates from human or mouse tissue. Corresponding PamChips® either analyze Serine-/Threonine kinases (STK-Chips) or Tyrosine kinases (PTK). Using suitable bioinformatics programs, differentially regulated kinases can be identified and, for example, specific targeting approaches can be tested in vitro.
See also: www.uke.de/english/departments-institutes/centers/university-cancer-center-hamburg-(ucch)%E2%80%94hubertus-wald-tumor-center/research/core-facilities/ucch-research-core-facilities-details.html
Functional kinome profiling. The PamGene's microarray assay for kinase activity profiling is based on measuring peptide phosphorylation by protein kinases. The PamChip®4 disposable consists of 4 identical arrays, each array containing up to 144 (STK) or 194 (PTK) peptides immobilized on a porous ceramic membrane. The peptide sequences (13 amino acids long) harbor phosphorylation sites derived from literature or computational predictions and are correlated with one or multiple upstream kinases. Fluorescently labelled anti-phospho antibodies are used to detect phosphorylation activity of kinases present in the sample. Images are later used by the BioNavigator® software to quantify the signals, generate heat maps and calculations and to perform the upstream kinase analysis, the prediction of relevant kinases. (With the license of PamGene Int. B.V.)
Project 1:
Signal transduction in head & neck cancer (HNSCC)
The aim of this project is to analyze the kinase-dependent signal transduction in head & neck cancer to identify tumor specific changes. Since we identified Src-family kinases (SFKs) to be frequently upregulated in head & neck cancer (Bussmann et al., 2021, Int J Cancer), following projects concentrate on the role of SFKs in head and neck cancer therapy resistance and how to target them (Thu Vu et al., 2023, Head Neck).
A recent project also focusses on FGF-receptors as drivers for oncogenesis and therapy resistance in a subset of head & neck cancer.
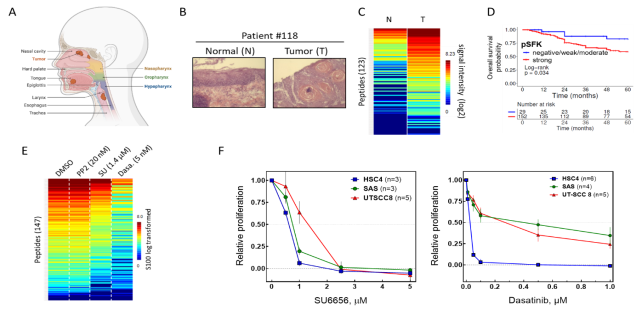
Signal transduction in HNSCC: From kinome profiling to molecular targeting. (A) Human cancer of the head and neck region (Biorender). (B) Human primary normal tissue and tumor samples were (C) analyzed using functional kinome profiling (see above) and kinases of the Src-family (SFK) were identified to be frequently upregulated in HNSCC. (D) SFK hyperactivity (increased SFK-autophosphorylation) is associated with worse prognosis in advanced HNSCC. (F) In SAS cells increased kinase activity can be blocked by SFK-inhibitor SU6656 and Abl/Src inhibitor Dasatinib. (F) Both inhibitos also block cell proliferation in head & neck tumor cell lines. (data from Bussmann et al., 2021, Int J Cancer & Thu Vu et al., 2023, Head Neck)
Most relevant Publications:
- Thu VuA, Akingunsade L, Hoffer K, Petersen C, Rothkamm K, Betz, C, Rieckmann T, Bußmann L, Kriegs M. Src family kinase (SFK) targeting in head and neck tumor cells using SU6656, PP2 and dasatinib. Head Neck. 2023 Jan;45(1):147-155.
- Bußmann L, Hoffer K, von Bargen CM, Droste C, Lange T, Kemmling J, Schröder-Schwarz J, Vu AT, Akingunsade L, Nollau P, Rangarajan S, de Wijn R, Oetting A, Müller C, Böckelmann LC, Zech HB, Berger JC, Möckelmann N, Busch CJ, Böttcher A, Gatzemeier F, Klinghammer K, Simnica D, Binder M, Struve N, Rieckmann T, Schumacher U, Clauditz TS, Betz CS, Petersen C, Rothkamm K, Münscher A, Kriegs M. Analyzing tyrosine kinase activity in head and neck cancer by functional kinomics: Identification of hyperactivated Src family kinases as prognostic markers and potential targets. Int J Cancer. 2021 Sep 1;149(5):1166-1180.
- Hintelmann K, Kriegs M, Rothkamm K, Rieckmann T. Improving the Efficacy of Tumor Radiosensitization Through Combined Molecular Targeting. Front Oncol. 2020, 10:1260. (review)
- Beizaei K, Gleißner L, Hoffer K, Bußmann L, Vu AT, Steinmeister L, Laban S, Möckelmann N, Münscher A, Petersen C, Rothkamm K, Kriegs M. Receptor tyrosine kinase MET as potential target of multi-kinase inhibitor and radiosensitizer sorafenib in HNSCC. Head Neck. 2019 Jan;41(1):208-215.
- Kriegs M, Clauditz T, Bußmann L, Hoffer K, Bartels J, Buhs S, Gerull H, Rieckmann T, Petersen C, Betz CS, Rothkamm K, Nollau P, Münscher A. Analyzing expression and phosphorylation of EGF-receptor in HNSCC. Sci Rep. 2019 Sep 19;9(1):13564.
- Braig F, Kriegs M, Voigtlaender M, Habel B, Grob T, Biskup K, Blanchard V, Sack M, Thalhammer A, Ben Batalla I, Braren I, Laban S, Danielczyk A, Goletz S, Jakubowicz E, Märkl B, Trepel M, Knecht R, Riecken K, Fehse B, Loges S, Bokemeyer C, Binder M. Cetuximab Resistance in Head and Neck Cancer Is Mediated by EGFR-K521 Polymorphism. Cancer Res, 2017 Mar 1;77(5):1188-1199.
- Kriegs M, Kasten-Pisula U, Riepen B, Hoffer K, Struve N, Myllynen L, Braig F, Binder M, Rieckmann T, Grénman R, Petersen C, Dikomey E, Rothkamm K. Radiosensitization of HNSCC cells by EGFR inhibition depends on the induction of cell cycle arrests. Oncotarget, 2016, 7(29):45122-45133.
- Mockelmann N, Rieckmann T, Busch CJ, Becker B, Gleissner L, Hoffer K, Omniczynski M, Steinmeister L, Laban S, Grenman R, Petersen C, Rothkamm K, Dikomey E, Knecht R, Kriegs M. Effect of sorafenib on cisplatin-based chemoradiation in head and neck cancer cells. Oncotarget, 2016, ;7(17):23542-51.
- Myllynen L*, Kwiatkowski M*, Gleißner L*, Riepen B, Hoffer K, Wurlitzer M, Petersen C, Dikomey E, Rothkamm K, Schlüter H**, Kriegs M**. *shared-first authorship; **shared-last authorship. Quantitative proteomics unveiled: Regulation of DNA double strand break repair by EGFR involves PARP1. Radiother Oncol, 2015,116(3):423-30.
Project 2:
Influence of kinase signaling on therapy sensitivity in Glioblastoma (GBM)
This project focuses on Glioblastoma multiforme (GBM), which is an aggressive primary brain tumor, characterized by rapid progression and oncogene activation. One common and frequently expressed oncogene in GBM is the constitutively activated epidermal growth factor receptor deletion mutant EGFRvIII. EGFRvIII represents a typical oncoprotein that activates different signaling pathways altering thereby multiple cellular processes. Therefore, understanding EGFRvIII-mediated signaling and analyzing how EGFRvIII impacts therapy relevant processes like DNA repair, replication, transcription and cellular signaling are main research interests. However, the influence of other kinases on the biology and radiation sensitivity of GBM is also the subject of our investigations. By understanding these complex processes, we hope to identify specific and new targeting approaches to individualize GBM therapy and to finally form the molecular rationale to improve overall survival.
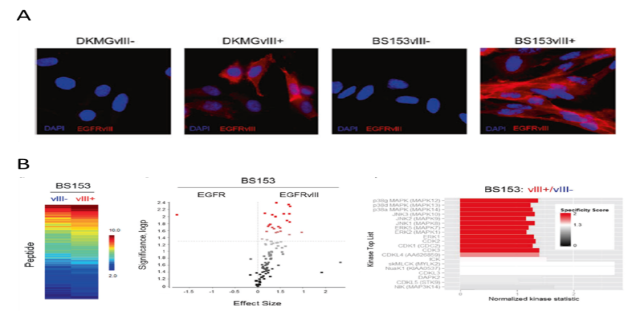
Signal transduction in GBM. (A) Using DKGM and BS153 cells lines we established a model system do analyze the EGFRvIII in GBM. Immunofluorescence staining of EGFRvIII (red) in sub-celllines with EGFRvIII expression (vIII+) and sub-celllines without EGFRvIII expression (vIII-), blue: DAPI stainig. (B) Functional kinome profling identifies upregulated MAP-Kinases in EGFRvIII+ GBM cells. (Heatmap; Two-group comparison of EGFRvIII+ vs. EGFRvIII− depicted as a volcano plot; Upstream kinase analysis of EGFRvIII+ vs. EGFRvIII−; top 20 of the differentially regulated kinases. Data from (Boyd, Struve et al., 2016, Nanoscale and Struve et al., 2020, Oncogene).
Most relevant Publications:
Jahnke K, Struve N, Hofmann D, Gote MJ, Bach M, Kriegs M*, Hausmann M*. Formation of EGFRwt/EGFRvIII homo- and hetero-dimers in glioblastoma cells as detected by single molecule localization microscopy. *shared-last authorship. Nanoscale. 2024 Aug 15;16(32):15240-15255.
Eckhardt A, Drexler R, Schoof M, Struve N, Capper D, Jelgersma C, Onken J, Harter PN, Weber KJ, Divé I, Rothkamm K, Hoffer K, Klumpp L, Ganser K, Petersen C, Ricklefs F, Kriegs M*, Schüller U*. *shared-last authorship. Mean global DNA methylation serves as independent prognostic marker in IDH-wildtype glioblastoma. Neuro Oncol. 2024 Mar 4;26(3):503-513.
Cetin MH, Rieckmann T, Hoffer K, Riepen B, Christiansen S, Gatzemeier F, Feyerabend S, Schoof M, Schüller U, Petersen C, Mynarek M, Rothkamm K, Kriegs M, Struve N. G2 checkpoint targeting via Wee1 inhibition radiosensitizes EGFRvIII-positive glioblastoma cells. Radiat Oncol. 2023 Jan 29;18(1):19.
Struve N, Hoffer K, Weik AS, Riepen B, Krug L, Cetin MH, Burmester J, Ott L, Liebing J, Gatzemeier F, Müller-Goebel J, Bußmann L, Parplys AC, Unger K, Mansour WY, Schüller U, Rieckmann T, Petersen C, Rothkamm K, Short SC*, Kriegs M*. *shared-last authorship. Increased replication stress and R-loop accumulation in EGFRvIII expressing glioblastoma present new therapeutic opportunities. Neuro-Oncology Advances. 04 December 2021.
Struve N, Binder ZA, Stead LF, Brend T, Bagley SJ, Faulkner C, Ott L, Müller-Goebel J, Weik AS, Hoffer K, Krug L, Rieckmann T, Bußmann L, Henze M, Morrissette JJD, Kurian KM, Schüller U, Petersen C, Rothkamm K, O Rourke DM, Short SC, Kriegs M. EGFRvIII upregulates DNA mismatch repair resulting in increased temozolomide sensitivity of MGMT promoter methylated glioblastoma. Oncogene. 2020 Apr;39(15):3041-3055. doi: 10.1038/s41388-020-1208-5.
Boyd PS*, Struve N*, Bach M, Eberle JP, Gote M, Schock F, Cremer C, Kriegs M**, Hausmann M**. *shared-first authorship; **shared-last authorship. Clustered localization of EGFRvIII in glioblastoma cells as detected by high precision localization microscopy. Nanoscale, 2016, 28;8(48):20037-20047.
Struve N, Riedel M, Schulte A, Rieckmann T, Grob TJ, Gal A, Rothkamm K, Lamszus K, Petersen C, Dikomey E, Kriegs M. EGFRvIII does not affect radiosensitivity with or without gefitinib treatment in glioblastoma cells. Oncotarget, 2015, 20;6(32):33867-77.
Project 3:
Radiation induced kinase signaling
Although it is already known, that X-rays can alter the kinase signaling in tumor cells, no systematic and functional studies have been performed so far. The aim of this project is to systematically analyze changes in the kinome of tumor cells induced by X-rays. For these studies we use primary tumor tissue cultures, so called ex vivo cultures (Zech et al., 2022, Radiother Oncol). We have shown that signal transduction is preserved in these cultures (Berger et al., 2021, Oral Oncol) while established tumor cells show significant differences in kinase signaling compared to primary tumor tissue.
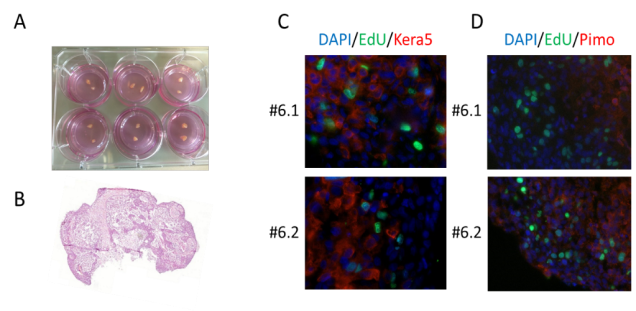
Ex vivo cultivation of head and neck tumors. (A) HNSCC biopsies were subdivided in 300 µm thick slices and cultured on Millicell® membranes. (B-D) Validation of morphology and vitality in HNSCC samples. Sample #6 is shown as a representative example. (B) H&E staining. (C) Detection of proliferating cells by immunofluorescence using EdU incorporation (green). DNA staining using DAPI (blue) served as a control. HNSCC cells were detected using a keratin 5-specific antibody (red). (D) Detection of hypoxia by immunofluorescence using pimonidazole (red). DNA staining using DAPI (blue) and EdU incorporation (green) served as controls. (Data from Berger et al., 2021, Oral Oncol)
Most relevant Publications:
Zech HB, Berger J, Mansour WY, Nordquist L, von Bargen C, Bußmann L, Oetting A, Christiansen S, Möckelmann N, Böttcher A, Busch CJ, Petersen C, Betz C, Rothkamm K, Kriegs M, Köcher S, Rieckmann T. Patient derived ex vivo tissue slice cultures demonstrate a profound DNA double-strand break repair defect in HPV-positive oropharyngeal head and neck cancer. Radiother Oncol. 2022 Jan 27:S0167-8140(22)00022-6.
Berger J, Zech HB, Hoffer K, von Bargen CM, Nordquist L, Bussmann L, Gatzemeier F, Busch CJ, Möckelmann N, Münscher A, Betz CS, Petersen C, Rothkamm K, Rieckmann T, Köcher S*, Kriegs M*. *shared-last authorship. Kinomic comparison of snap frozen and ex vivo-cultured head and neck tumors. Oral Oncol. 2021 Dec;123:105603.
Project 4:
Predictive marker for immune checkpoint inhibition
The aim of this project is to identify responders and non-responders to immune checkpoint inhibition (ICI) to guide further therapy of head and neck squamous cell carcinoma (HNSCC) patients.
Immune checkpoint inhibitors have already been approved as 1st and 2nd line therapy of relapsed or distant metastatic head and neck squamous cell carcinoma (R/M-HNSCC). However, only 13-18% respond to therapy with no definitive predictive markers available resulting in a great need to establish predictive tests identifying responding and non-responding patients.
For ICI therapy of advanced malignant melanoma and non small cell lung carcinoma it has been demonstrated, that specific kinase activity clusters of peripheral blood mononuclear cells (PBMC) can be predictive. These clusters result from functional kinome profiling and might also be promising in R/M-HNSCC. To test this, liquid biopsies are collected from HNSCC patients before and under ICI treatment. PBMC are isolated and functional kinome profiling is performed.
This method may also allow to monitor the patients` individual response during ICI treatment enabling the early detection of a progresses. Such a predictive test would support to identify responding versus non-responding patients and to guide non-responders to a more appropriate therapy. This would also enable a more efficient economical use of the expensive therapeutics.
Project 5:
Response prediction of kinase inhibitors in personalized cancer therapy
Based on our experience from the previous projects, our goal is to use kinome profiling in the personalized therapy of cancer patients. Together with colleagues from the UCCH molecular tumorboard, the Department of Oncology & Hematology (II. Med.) and the Department of Otorhinolaryngology we use functional kinome profiling to identify hyperactive kianses in tumors with no targetable genetic mutations or predict the response to kinase inhibitors by analyzing their in vitro effect on kinase activity in primary tumor samples. The transfer of our pre-clinical experience for clinical applications is the central task for our future research.
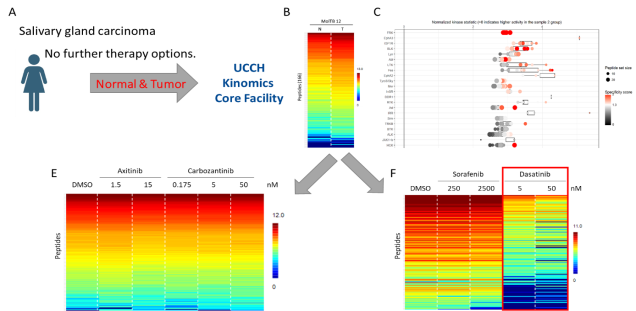
Potential use of kinome profiling in personalized medicine. (A, B) Normal and tumor tissue from a patient with advanced salivary gland carcinoma was analyzed by functional kinome profiling. (C) Several tyrsosine kinases were identified to be upregulated in the tumor sample. (E, F) Based on these results, different kinase inhibitors were tested in vitro (F) with only dasatinib showing a significant inhibitory potential.
Grants: Federal Ministry of Education and Research (BMBF); Hamburger Krebsgesellschaft; Wilhelm Sander-Stiftung; Werner Otto-Stiftung; Roggenbuck-Stiftung; Stiftung Tumorforschung Kopf-Hals; Brigitte & Dr. Konstanze Wegener-Stiftung Hamburger Stiftung zur Förderung der Krebsbekämpfung; Forschungsförderungsfonds des Fachbereichs Medizin; Spierling-Stiftung; Landesforschungsförderung der Hansestadt Hamburg
6. Fellowships & Awards:
2025 UCCH Clinician Scientist (Ramke)
2024 DeGBS Best Paper of the Year (Eckhardt et al., 2024, Neuro Oncol)
2024 UCCH Clinician Scientist (Röhrle)
2020 Research award of the Hamburger Cancer Society (Struve/Kriegs)
2020 Hubertus Wald Junior Investigator Award 2020 (Struve)
2020 MSNZ Clinician Scientist (Bußmann)
2019 MD stipend E.W. Kuhlmann-Stiftung (Cetin)
2018 UCCH Clinician Scientist (Berger, alumna)
2017 UCCH Clinician Scientist (Bußmann)
2016 MD stipend Werner Otto-Stiftung (Struve/Müller-Goebel, alumnus)
2016 Friedrich Zywitz-Promotionspreis (Struve)
2016 UCCH Clinician Scientist (Bußmann)
2015 UKE International Travel Scholarships (Kriegs/Struve)
2015 GlaxoSmithKline Stiftung Reisestipendium (Struve)
2015 Oncoray Poster Prize (Gleißner, alumna)
2014 Friedrich Zywitz-Promotionspreis (Myllynen, alumna)
2013 UCCH Clinician Scientist (Möckelmann, alumnus)
2012 UCCH Clinician Scientist (Busch, alumna)
2011 UCCH Clinician Scientist (Laban, alumnus)